Benchmark 1. MCA lung dataset annotation using ref_tabula_muris_drop reference
library(clustifyr) library(clustifyrdata) l_mat <- clustifyrdata::MCA_lung_mat l_meta <- clustifyrdata::MCA_lung_meta # find lung references, remove generic terms lung_cols <- grep("-Lung", colnames(ref_tabula_muris_drop), value = TRUE) tml_ref <- ref_tabula_muris_drop[, lung_cols] tml_ref <- tml_ref[, -c(8, 13)] # default with all genes start <- proc.time() res <- clustify( input = l_mat, ref_mat = tml_ref, metadata = l_meta, cluster_col = "Annotation" ) #> [1] "use" res_allgenes <- cor_to_call( cor_mat = res, metadata = l_meta, cluster_col = "Annotation" ) end <- proc.time() names(res_allgenes) <- c("MCA annotation", "clustifyr call", "r") print(end - start) #> user system elapsed #> 1.962 0.468 2.439 print(res_allgenes, n = nrow(res_allgenes)) #> # A tibble: 32 x 3 #> # Groups: Annotation [32] #> `MCA annotation` `clustifyr call` r #> <chr> <chr> <dbl> #> 1 Alveolar macrophage_Ear2 high(… alveolar macrophage-Lung 0.878 #> 2 Alveolar macrophage_Pclaf high… alveolar macrophage-Lung 0.714 #> 3 B Cell(Lung) B cell-Lung 0.836 #> 4 Ig−producing B cell(Lung) B cell-Lung 0.577 #> 5 Ciliated cell(Lung) ciliated columnar cell of tracheobronc… 0.820 #> 6 Plasmacytoid dendritic cell(Lu… classical monocyte-Lung-CLASH! 0.847 #> 7 Eosinophil granulocyte(Lung) leukocyte-Lung 0.716 #> 8 Neutrophil granulocyte(Lung) leukocyte-Lung 0.634 #> 9 Endothelial cell_Kdr high(Lung) lung endothelial cell-Lung 0.747 #> 10 Endothelial cell_Tmem100 high(… lung endothelial cell-Lung 0.803 #> 11 Endothelial cells_Vwf high(Lun… lung endothelial cell-Lung 0.764 #> 12 Basophil(Lung) mast cell-Lung 0.440 #> 13 NK Cell(Lung) natural killer cell-Lung 0.804 #> 14 Conventional dendritic cell_Gn… non-classical monocyte-Lung-CLASH! 0.789 #> 15 Stromal cell_Acta2 high(Lung) stromal cell-Lung 0.646 #> 16 Stromal cell_Dcn high(Lung) stromal cell-Lung 0.814 #> 17 Stromal cell_Inmt high(Lung) stromal cell-Lung 0.817 #> 18 Dividing T cells(Lung) T cell-Lung 0.720 #> 19 Nuocyte(Lung) T cell-Lung 0.758 #> 20 T Cell_Cd8b1 high(Lung) T cell-Lung 0.826 #> 21 Alveolar bipotent progenitor(L… alveolar epithelial type 2 cells-Lung 0.663 #> 22 AT1 Cell(Lung) alveolar epithelial type 2 cells-Lung 0.770 #> 23 AT2 Cell(Lung) alveolar epithelial type 2 cells-Lung 0.880 #> 24 Clara Cell(Lung) alveolar epithelial type 2 cells-Lung 0.733 #> 25 Dividing cells(Lung) alveolar epithelial type 2 cells-Lung 0.647 #> 26 Conventional dendritic cell_H2… dendritic cells and interstital macrop… 0.550 #> 27 Conventional dendritic cell_Mg… dendritic cells and interstital macrop… 0.788 #> 28 Conventional dendritic cell_Tu… dendritic cells and interstital macrop… 0.671 #> 29 Dendritic cell_Naaa high(Lung) dendritic cells and interstital macrop… 0.802 #> 30 Dividing dendritic cells(Lung) dendritic cells and interstital macrop… 0.676 #> 31 Interstitial macrophage(Lung) dendritic cells and interstital macrop… 0.804 #> 32 Monocyte progenitor cell(Lung) dendritic cells and interstital macrop… 0.581
benchmark 2. Using sorted microarray data to classify 10x PBMC example data, available in clustifyrdata package
full_pbmc_matrix <- clustifyrdata::pbmc_matrix full_pbmc_meta <- clustifyrdata::pbmc_meta microarray_ref <- clustifyrdata::ref_hema_microarray start <- proc.time() res <- clustify( input = full_pbmc_matrix, ref_mat = microarray_ref, metadata = full_pbmc_meta, query_genes = pbmc_vargenes[1:500], cluster_col = "classified" ) #> [1] "use" res2 <- cor_to_call(res, threshold = 0.5) end <- proc.time() names(res2) <- c("manual annotation", "clustifyr call", "r") print(end - start) #> user system elapsed #> 0.087 0.005 0.093 print(res2, n = nrow(res2)) #> # A tibble: 9 x 3 #> # Groups: cluster [9] #> `manual annotation` `clustifyr call` r #> <chr> <chr> <dbl> #> 1 Memory CD4 T CD4+ Effector Memory 0.585 #> 2 Naive CD4 T CD4+ Effector Memory 0.594 #> 3 CD8 T CD8+ Effector Memory 0.602 #> 4 NK Mature NK cell_CD56+ CD16+ CD3- 0.537 #> 5 Platelet unassigned 0.298 #> 6 CD14+ Mono Monocyte 0.593 #> 7 FCGR3A+ Mono Monocyte 0.559 #> 8 DC Myeloid Dendritic Cell 0.556 #> 9 B Naïve B-cells 0.634
- Please see manuscript for full benchmarking.
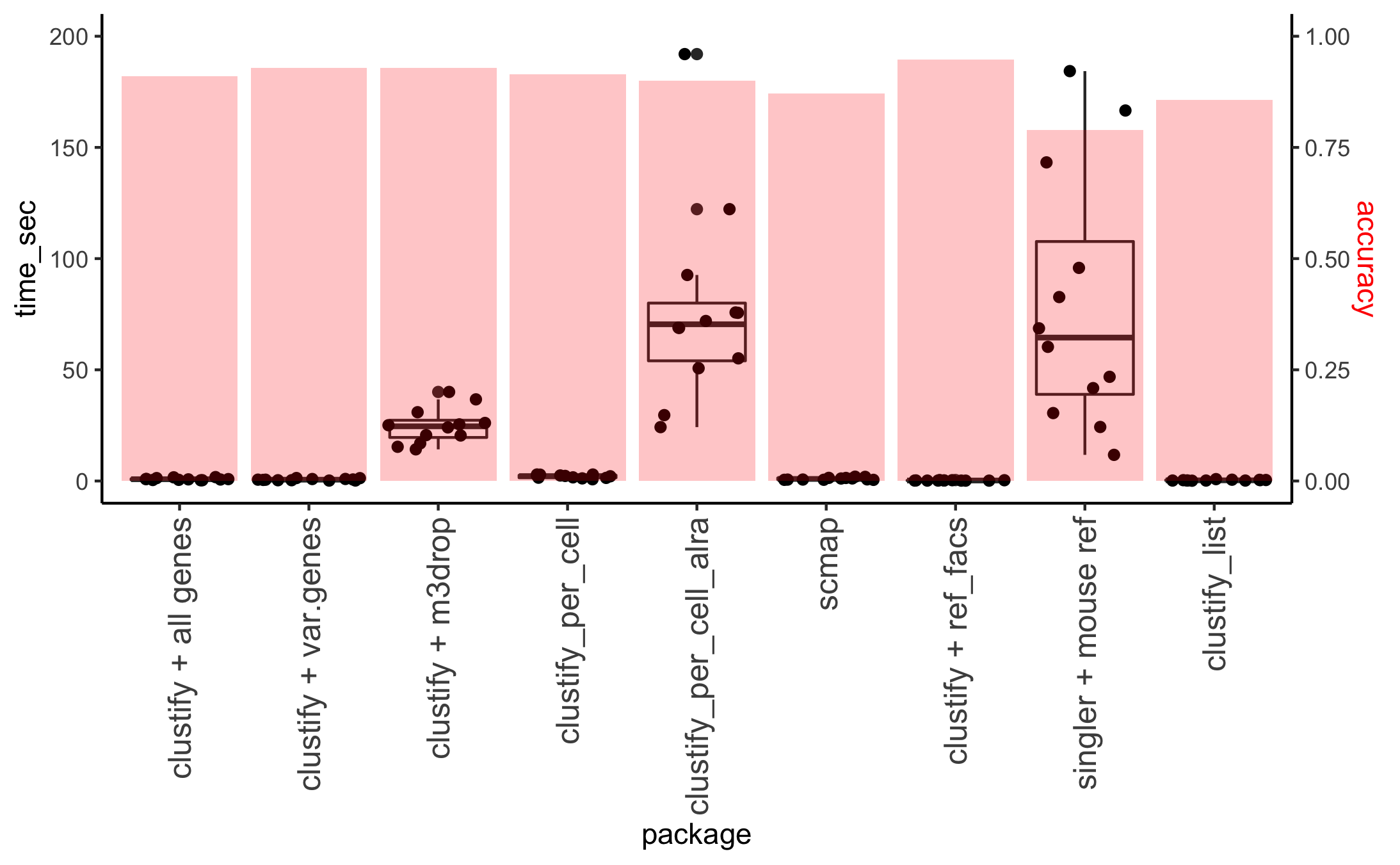
Comparison with other methods
using Tablua Muris (drop and facs samples) 12 shared tissues, which can be downloaded as seurat objects
- Building reference and then mapping:
default clustify, with all genes
clustify, pulling var.genes from seurat objects
clustify, using M3Drop for feature selection
clustify, using per_cell = TRUE option, and then assign cluster consensus ident with collapse_to_cluster = TRUE
clustify, after ALRA imputation, using per_cell = TRUE option, and then assign cluster consensus ident with collapse_to_cluster = TRUE
scmap-cluster
- Mapping from prebuilt all-encompassing references to the drop samples:
clustify, using ref_tabula_muris_facs
singleR, using default built-in mouse references without fine tuning
- Generate marker gene list (of 30 genes per reference identity), and then mapping
default clustify_list