single cell RNA-seq workshop
R installation
Running R/Rstudio with docker
In this post we will discuss how to use docker to run R and Rstudio in a standardized environment.
Working with your own single cell data
Guidelines for datasets to work with in the workshop, information on where to get public datasets, and examples of how to load different data formats into R.
Prerequisite material
Interacting with R in Rstudio
A tutorial for navigating the Rstudio IDE, RMarkdown, and setting up Rstudio projects to organize each class.
Intro_scRNAseq_QC_PCA
The goal of this lecture is to give an overview of scRNA seq, quality control, filtering, and briefly touch on PCA
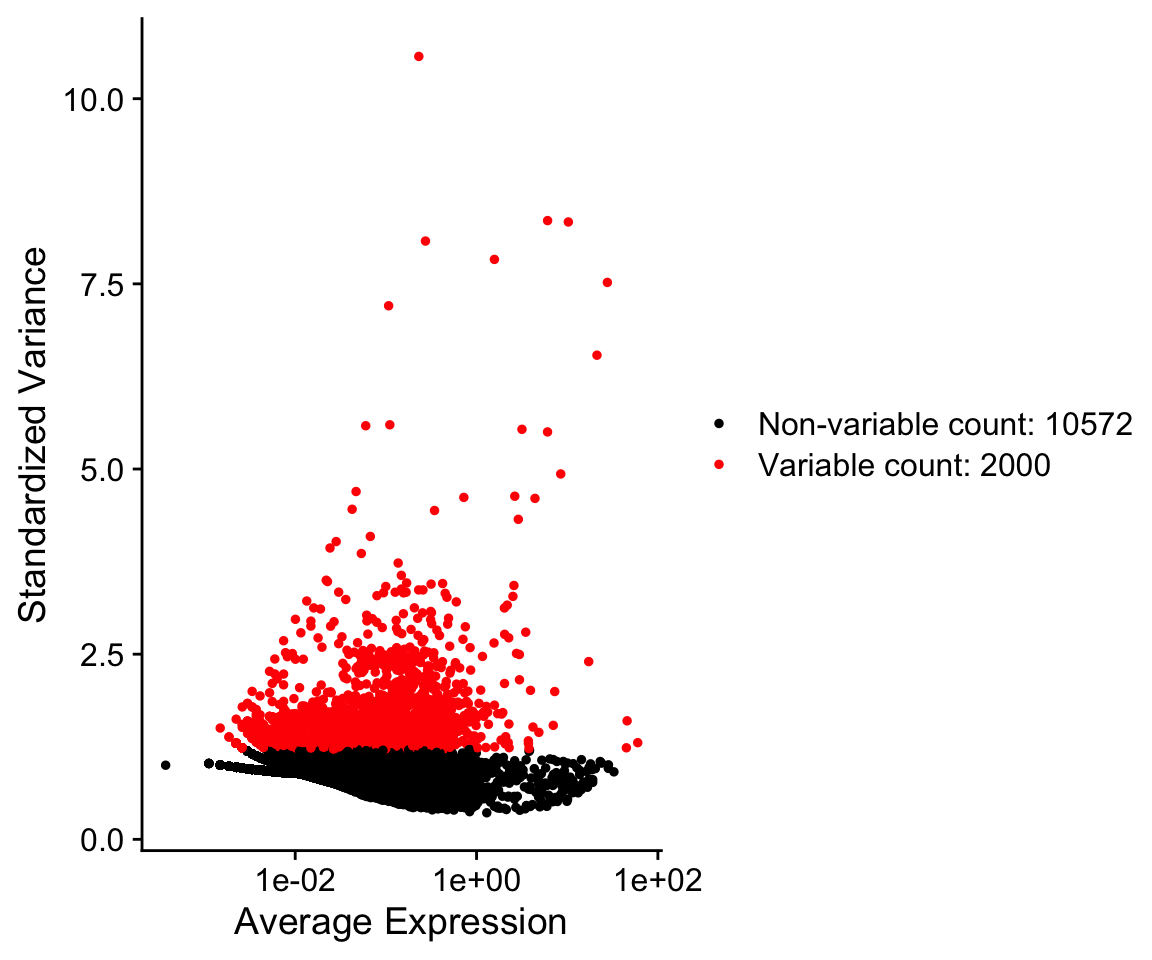
Class 2: Dimensionality reduction, clustering, differential expression, cell type annotation
In this class we will discuss scRNA-seq data processing for visualization and defining transcriptome features.
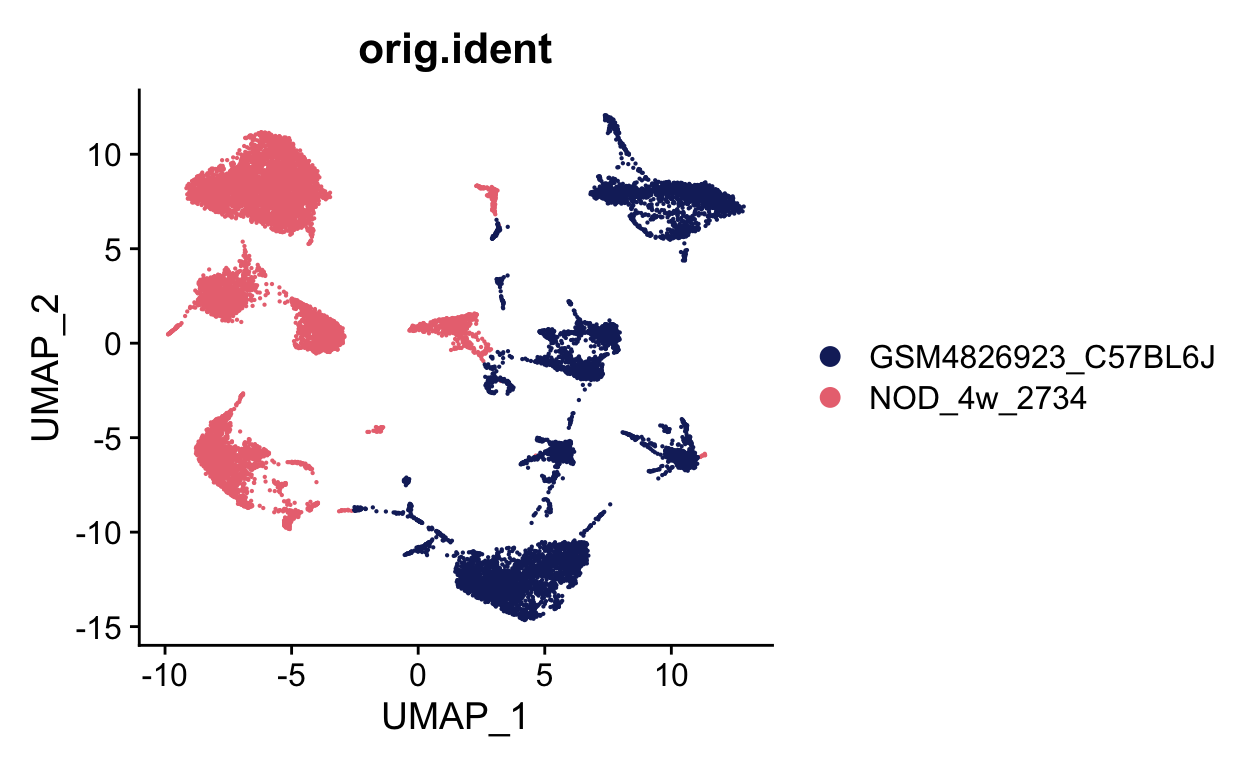
Class 3: Batch correction and pseudotime
This tutorial walks through batch correction between two samples from different groups with some overlapping cell types using harmony. In the second half of the tutorial, we use timecourse data to run and evaluate pseudotime using slingshot.
Class 4: Multi-modal data
This tutorial describes a basic CITE-seq analysis workflow.

FAQ
Answers to common questions