We’ve built a docker image that contains the R packages used in the workshop and made it available on dockerhub. This image can be used if you have issues installing the recommended R packages.
What is docker?
Docker is an application that allows identical software environments (called images) to be run in different operating systems (MacOS, Linux or Windows). The standardized software environment (programs, data, and configuration) takes away the headache of trying to install and maintain software. Practically using docker also enforces a reproducible analysis environment.
In this post we will discuss the basics of how to:
- install docker
- run docker on the command-line/terminal
- run a docker container containing R, Rstudio
- run a docker container for this workshop that contains a variety of single cell packages
Install docker
MacOS: Install Docker desktop here: https://docs.docker.com/desktop/mac/install/
Windows: If you do not have the WSL enabled or installed, following the instructions here: https://docs.microsoft.com/en-us/windows/wsl/install
Then download and install Docker desktop for windows: https://docs.docker.com/desktop/windows/install/
Open the docker application to confirm installation.
How to run a docker container
A docker image refers to the standarized software environment. A docker container is the copy of the image that is downloaded and run on a specific computer.
If the docker application is running, then you should be able to run a docker container in the command line. Open up Terminal in macOS or PowerShell in windows.
If you run the following command, an image with R and Rstudio installed (rocker/tidyverse) will be downloaded from dockerhub and activated.
docker run --rm -e PASSWORD=rna -p 8787:8787 rocker/tidyverseWe’ll explain the other command arguments in a moment.
You’ll see some messages that will look something like this:
Unable to find image 'rocker/tidyverse:latest' locally
latest: Pulling from rocker/tidyverse
...
...
Status: Downloaded newer image for rocker/verse:latest
[s6-init] making user provided files available at /var/run/s6/etc...exited 0.
[s6-init] ensuring user provided files have correct perms...exited 0.
[fix-attrs.d] applying ownership & permissions fixes...
[fix-attrs.d] done.
[cont-init.d] executing container initialization scripts...
[cont-init.d] userconf: executing...
skipping /var/run/s6/container_environment/HOME
skipping /var/run/s6/container_environment/PASSWORD
skipping /var/run/s6/container_environment/RSTUDIO_VERSION
[cont-init.d] userconf: exited 0.
[cont-init.d] done.
[services.d] starting services
[services.d] done.After running this command an rstudio instance will be running on your computer that you can access through your web browser. Open a browser and enter http://, followed by your ip address, followed by :8787. In you are on a linux or MacOS machine you can navigate to http//localhost:8787. If you do not know the IP address you can find it in a few ways.
- If you select the container in the docker desktop app, you can directly open the container in a browser by clicking the
open in browserbutton.
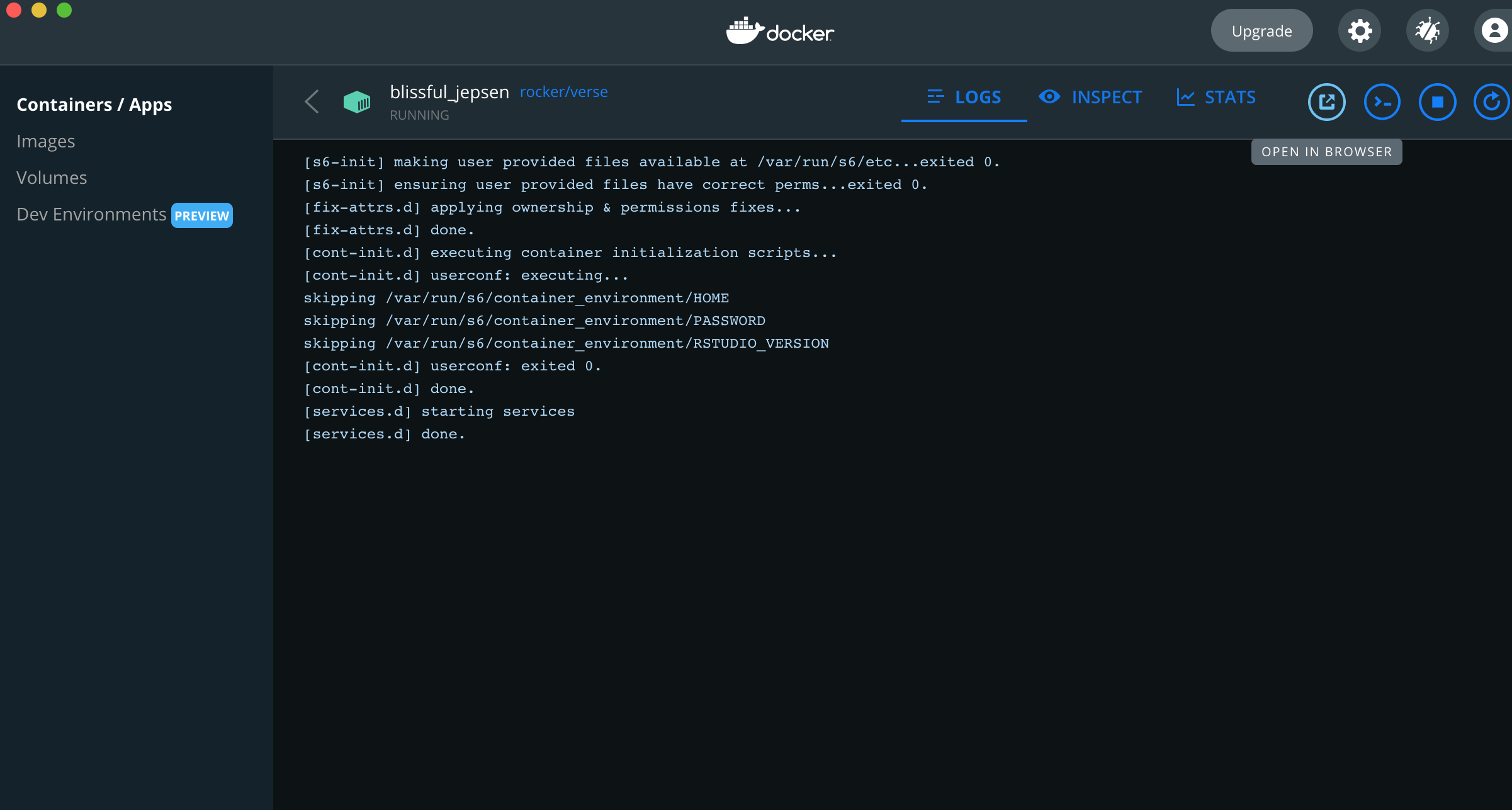
- You can obtain the IP address from the command line.
Get list of all of your containers using docker ps
docker ps
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
d2629b78er4f rocker/verse "/init" 14 minutes ago Up 14 minutes 0.0.0.0:8787->8787/tcp, :::8787->8787/tcp blissful_jepsenThen use docker inspect get ip address using the value found under “CONTAINER ID”
docker inspect d2629b78er4fJust get the ip-address
docker inspect --format='{{range .NetworkSettings.Networks}}{{.IPAddress}}{{end}}' d2629b78er4fNavigate to http://Your.I.p.Address:8787
You should see a Rstudio login page
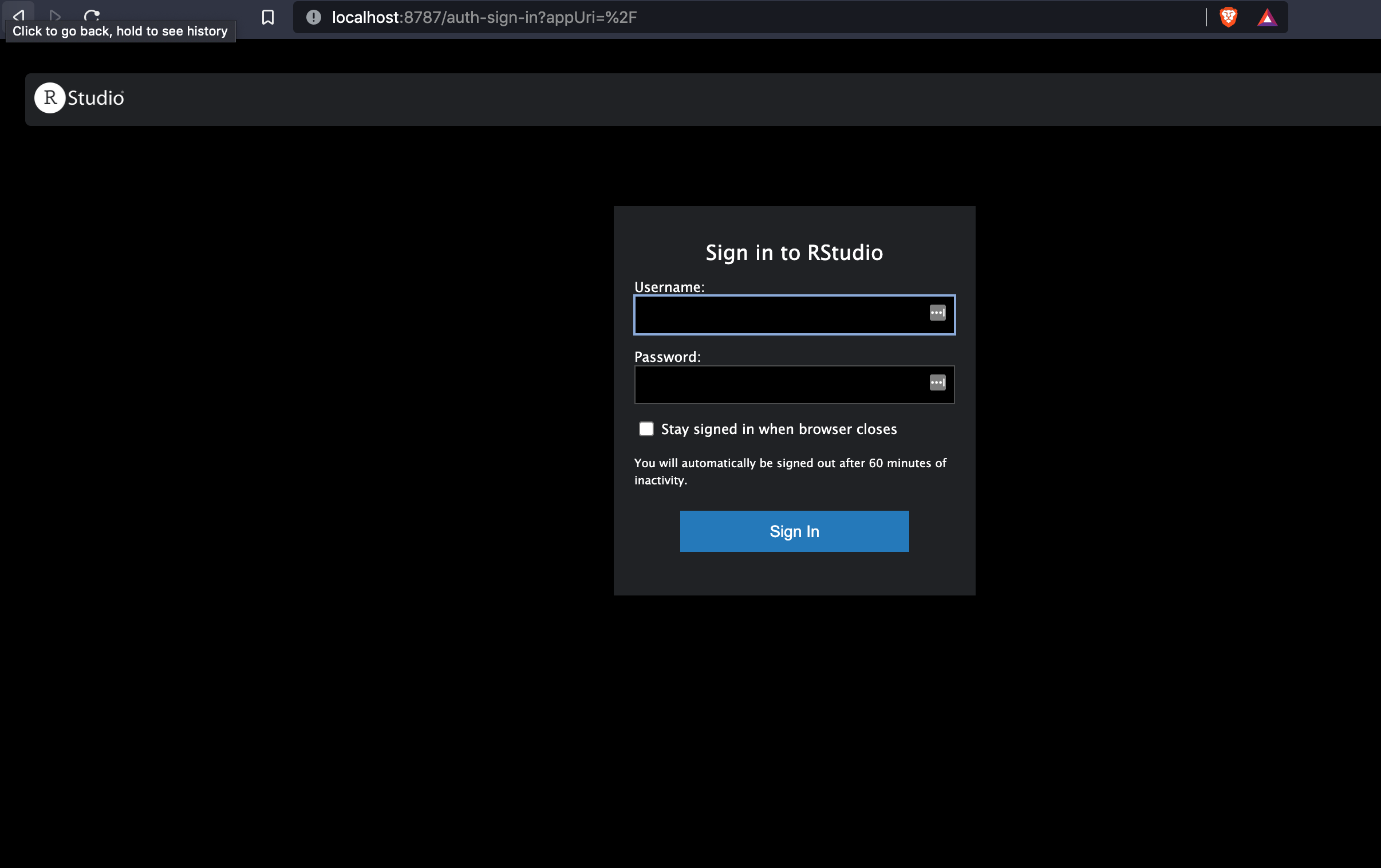
Login to rstudio with username rstudio and password rna. You should see Rstudio running in your browser.
Once you are done playing around in rstudio you can stop the container by terminating the process in your terminal:
Go back to your terminal and run CTRL + C in macOS or linux, or Ctrl+Break in Windows
You’ll see a few more messages, then the command will exit back to your prompt
s6-finish] waiting for services.
[s6-finish] sending all processes the TERM signal.
[s6-finish] sending all processes the KILL signal and exiting.
$When you initiated the docker container we used the following command:
docker run --rm -e PASSWORD=rna -p 8787:8787 rocker/tidyverseThe --rm is a special flag that tells docker to delete the container after exiting. You can verify this occurred by looking at your containers in Docker Desktop or by runnning docker ps on the command line.
The --rm option is a good way to start working with docker. If you don’t include this a new container will be made each time you execute docker run unless you provide different flags. This can quickly take up disk space if you are not careful so we recommend that you use --rm for now. The other advantage is this approach ensures that your R environment is restored to a clean environment each time you start the container.
The -e PASSWORD=rna parameter specified that the password for logging into rstudio should be rna.
The -p 8787:8787 parameter specified that Rstudio should be served on the 8787 port.
How to use and create files on your local computer
You may have noticed that you do not have access to local files in the rstudio instance by default. This is expected behavior as the docker container exists in an isolated environment for your local computer.
If you want to provide rstudio access to a local directory to allow reading and writing local files do the following:
Find the path to a directory on your computer e.g. for the desktop on macOs use
~/Desktopor for documents in windowsC:\Documentsadd the
-vparameter which has the syntax-v /path/to/local/directory:/path/in/container. Because we are usingrstudio, if we put files into the container at/home/rstudio, they will be visible to rstudio.
Try it out: Here I am making my a class directory on my desktop (on a macOS) visible to rstudio. The class directory has 1 file called hello-world.txt.
docker run --rm -v ~/Desktop/class:/home/rstudio -e PASSWORD=rna -p 8787:8787 rocker/tidyverseIn Rstudio I can now see the hello-world.txt file, and if I make a new file in R called docker-file.txt, it will now be visible on my local computer, and persist after I exit docker.
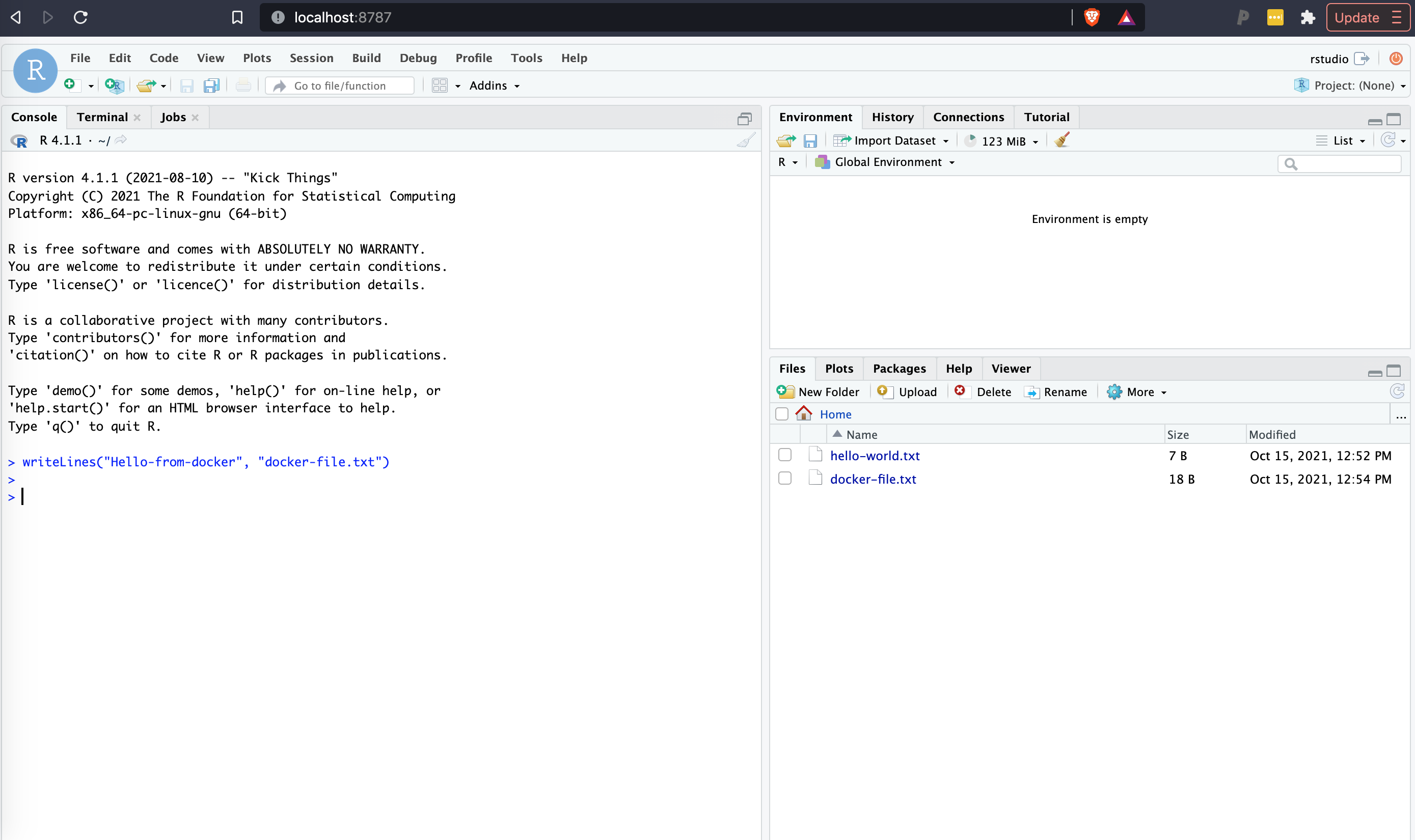
Similarly you could write a R script or Rmarkdown document, run it in rstudio, then save the script into your local files for future reuse.
Using docker for workshop
We have a docker image available on dockerhub that includes the packages that will be used in the class.
You can use this image in the same manner as above, instead using kenter/rstudio-scrnaseq:v0.2.
Try it out:
docker run --rm -e PASSWORD=rna -p 8787:8787 kenter/rstudio-scrnaseq:v0.2When you open rstudio you should be able to load Seurat and other single cell packages. We’ve also included a test-install.Rmd document that lists and loads all of the relevant installed packages, and tests basic Seurat commands.
Additional resources
If you’ve never used docker here are some useful tutorials on using docker:
https://bioconductor.org/help/docker/#quickstart
https://jsta.github.io/r-docker-tutorial/
https://replikation.github.io/bioinformatics_side/docker/docker/#important-commands