Out-of-bounds intervals are removed by default.
See also
https://bedtools.readthedocs.io/en/latest/content/tools/shift.html
Other single set operations:
bed_cluster(),
bed_complement(),
bed_flank(),
bed_genomecov(),
bed_merge(),
bed_partition(),
bed_slop()
Examples
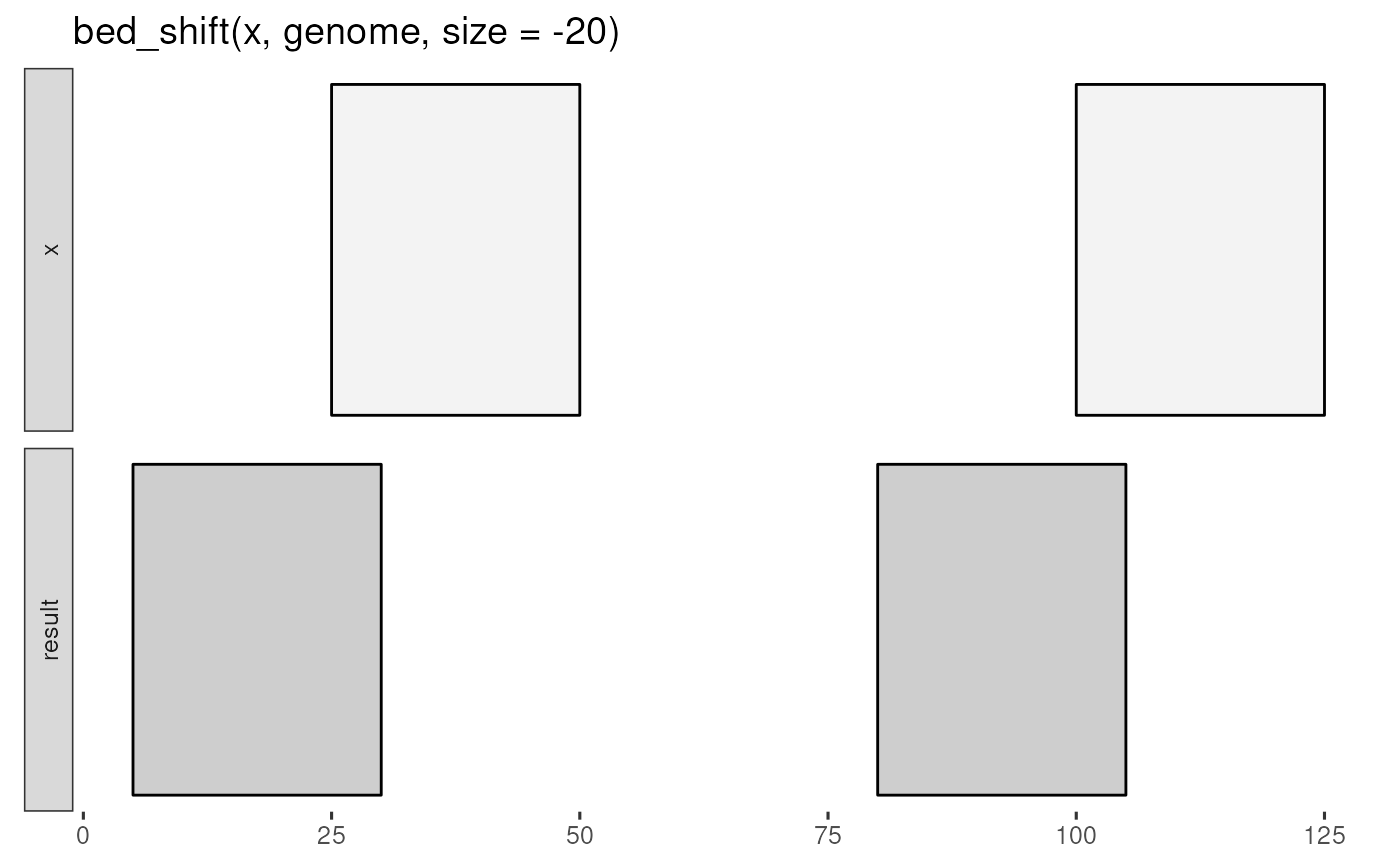
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 25, 50,
"chr1", 100, 125
)
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 125
)
bed_glyph(bed_shift(x, genome, size = -20))
 x <- tibble::tribble(
~chrom, ~start, ~end, ~strand,
"chr1", 100, 150, "+",
"chr1", 200, 250, "+",
"chr2", 300, 350, "+",
"chr2", 400, 450, "-",
"chr3", 500, 550, "-",
"chr3", 600, 650, "-"
)
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 1000,
"chr2", 2000,
"chr3", 3000
)
bed_shift(x, genome, 100)
#> # A tibble: 6 × 4
#> chrom start end strand
#> <chr> <dbl> <dbl> <chr>
#> 1 chr1 200 250 +
#> 2 chr1 300 350 +
#> 3 chr2 400 450 +
#> 4 chr2 500 550 -
#> 5 chr3 600 650 -
#> 6 chr3 700 750 -
bed_shift(x, genome, fraction = 0.5)
#> # A tibble: 6 × 4
#> chrom start end strand
#> <chr> <dbl> <dbl> <chr>
#> 1 chr1 125 175 +
#> 2 chr1 225 275 +
#> 3 chr2 325 375 +
#> 4 chr2 425 475 -
#> 5 chr3 525 575 -
#> 6 chr3 625 675 -
# shift with respect to strand
stranded <- dplyr::group_by(x, strand)
bed_shift(stranded, genome, 100)
#> # A tibble: 6 × 4
#> chrom start end strand
#> <chr> <dbl> <dbl> <chr>
#> 1 chr1 200 250 +
#> 2 chr1 300 350 +
#> 3 chr2 400 450 +
#> 4 chr2 300 350 -
#> 5 chr3 400 450 -
#> 6 chr3 500 550 -
x <- tibble::tribble(
~chrom, ~start, ~end, ~strand,
"chr1", 100, 150, "+",
"chr1", 200, 250, "+",
"chr2", 300, 350, "+",
"chr2", 400, 450, "-",
"chr3", 500, 550, "-",
"chr3", 600, 650, "-"
)
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 1000,
"chr2", 2000,
"chr3", 3000
)
bed_shift(x, genome, 100)
#> # A tibble: 6 × 4
#> chrom start end strand
#> <chr> <dbl> <dbl> <chr>
#> 1 chr1 200 250 +
#> 2 chr1 300 350 +
#> 3 chr2 400 450 +
#> 4 chr2 500 550 -
#> 5 chr3 600 650 -
#> 6 chr3 700 750 -
bed_shift(x, genome, fraction = 0.5)
#> # A tibble: 6 × 4
#> chrom start end strand
#> <chr> <dbl> <dbl> <chr>
#> 1 chr1 125 175 +
#> 2 chr1 225 275 +
#> 3 chr2 325 375 +
#> 4 chr2 425 475 -
#> 5 chr3 525 575 -
#> 6 chr3 625 675 -
# shift with respect to strand
stranded <- dplyr::group_by(x, strand)
bed_shift(stranded, genome, 100)
#> # A tibble: 6 × 4
#> chrom start end strand
#> <chr> <dbl> <dbl> <chr>
#> 1 chr1 200 250 +
#> 2 chr1 300 350 +
#> 3 chr2 400 450 +
#> 4 chr2 300 350 -
#> 5 chr3 400 450 -
#> 6 chr3 500 550 -
