Identify intervals in a genome not covered by a query.
See also
Other single set operations:
bed_cluster(),
bed_flank(),
bed_genomecov(),
bed_merge(),
bed_partition(),
bed_shift(),
bed_slop()
Examples
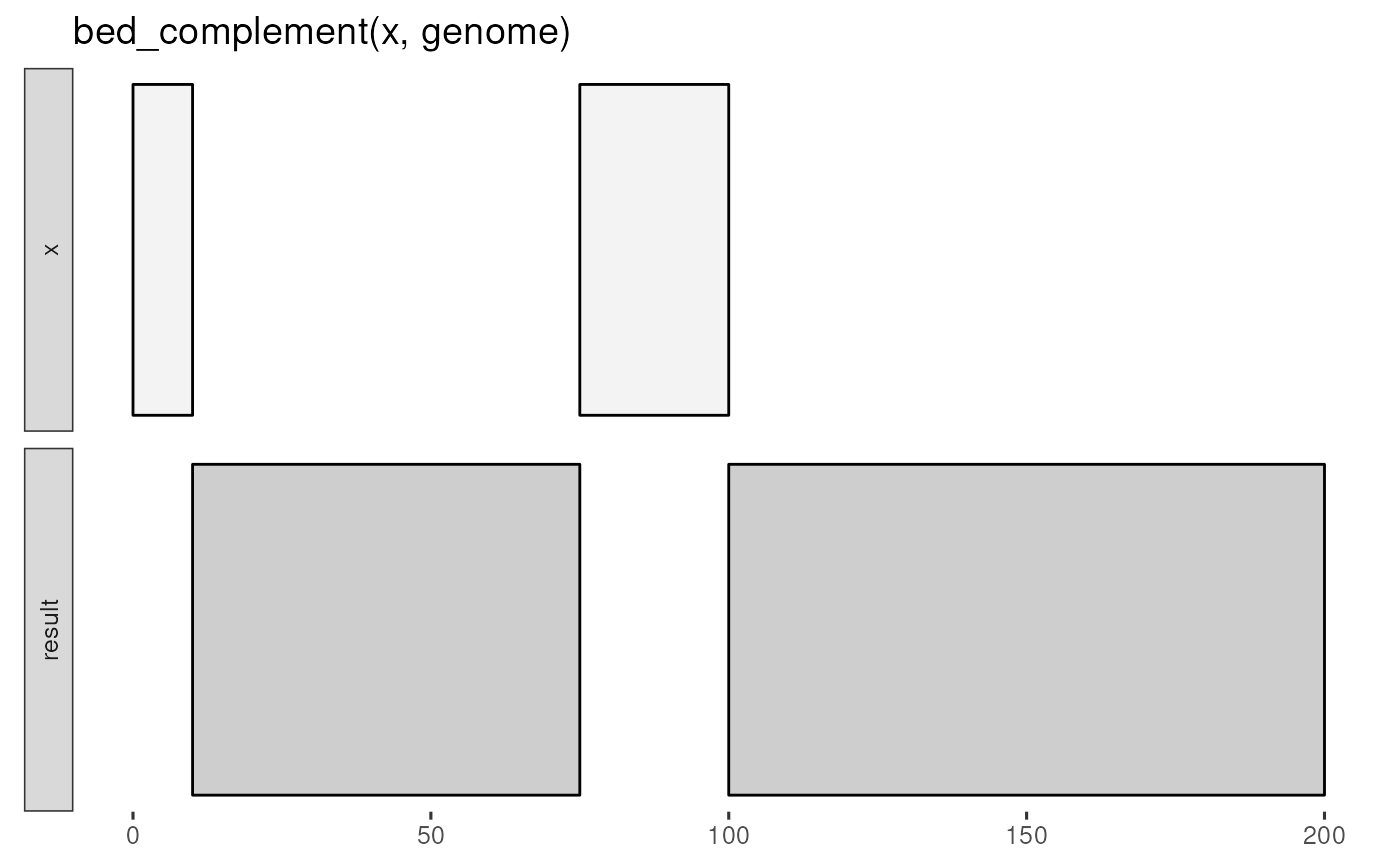
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 0, 10,
"chr1", 75, 100
)
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 200
)
bed_glyph(bed_complement(x, genome))
 genome <- tibble::tribble(
~chrom, ~size,
"chr1", 500,
"chr2", 600,
"chr3", 800
)
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 100, 300,
"chr1", 200, 400,
"chr2", 0, 100,
"chr2", 200, 400,
"chr3", 500, 600
)
# intervals not covered by x
bed_complement(x, genome)
#> # A tibble: 6 × 3
#> chrom start end
#> <chr> <dbl> <dbl>
#> 1 chr1 0 100
#> 2 chr1 400 500
#> 3 chr2 100 200
#> 4 chr2 400 600
#> 5 chr3 0 500
#> 6 chr3 600 800
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 500,
"chr2", 600,
"chr3", 800
)
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 100, 300,
"chr1", 200, 400,
"chr2", 0, 100,
"chr2", 200, 400,
"chr3", 500, 600
)
# intervals not covered by x
bed_complement(x, genome)
#> # A tibble: 6 × 3
#> chrom start end
#> <chr> <dbl> <dbl>
#> 1 chr1 0 100
#> 2 chr1 400 500
#> 3 chr2 100 200
#> 4 chr2 400 600
#> 5 chr3 0 500
#> 6 chr3 600 800
