Plot repertoire similarity
Usage
plot_similarity(
input,
data_col,
cluster_col,
group_col = NULL,
method = abdiv::jaccard,
chain = NULL,
chain_col = global$chain_col,
cluster_heatmap = TRUE,
sep = global$sep,
plot_colors = NULL,
plot_lvls = names(plot_colors),
rotate_labels = FALSE,
remove_upper_triangle = FALSE,
remove_diagonal = remove_upper_triangle,
...
)Arguments
- input
Single cell object or data.frame containing V(D)J data. If a data.frame is provided, the cell barcodes should be stored as row names.
- data_col
meta.data column containing values to use for calculating pairwise similarity between clusters, e.g. 'clonotype_id'
- cluster_col
meta.data column containing cluster IDs to use for calculating overlap
- group_col
meta.data column to use for grouping cluster IDs present in cluster_col. This is useful when there are multiple replicates or patients for each treatment condition.
- method
Method to use for comparing clusters, possible values are:
A function that takes two numeric vectors containing counts for each clonotype in the object, such as most beta diversity functions provided by the abdiv package. This will generate a heatmap.
'count', count the number of clonotypes overlapping between each cluster, this will generate a heatmap.
'circos', create a circos plot summarizing the overlap between clusters
- chain
Chain to use for comparing clusters. To perform calculations using a single chain, the column passed to the data_col argument must contain per-chain data such as CDR3 sequences. Set to NULL to include all chains.
- chain_col
meta.data column containing chains for each cell
- cluster_heatmap
If FALSE, rows and columns of heatmap will not be clustered.
- sep
Separator used for storing per-chain V(D)J data for each cell
- plot_colors
Character vector containing colors for plotting
- plot_lvls
Levels to use for ordering clusters
- rotate_labels
Should labels on circos plot be rotated to reduce overlapping text
- remove_upper_triangle
If TRUE, upper triangle for heatmap will not be shown.
- remove_diagonal
If TRUE, diagonal for heatmap will not be shown.
- ...
Additional arguments to pass to plotting function,
ComplexHeatmap::Heatmap()for heatmap,circlize::chordDiagram()for circos plot
Examples
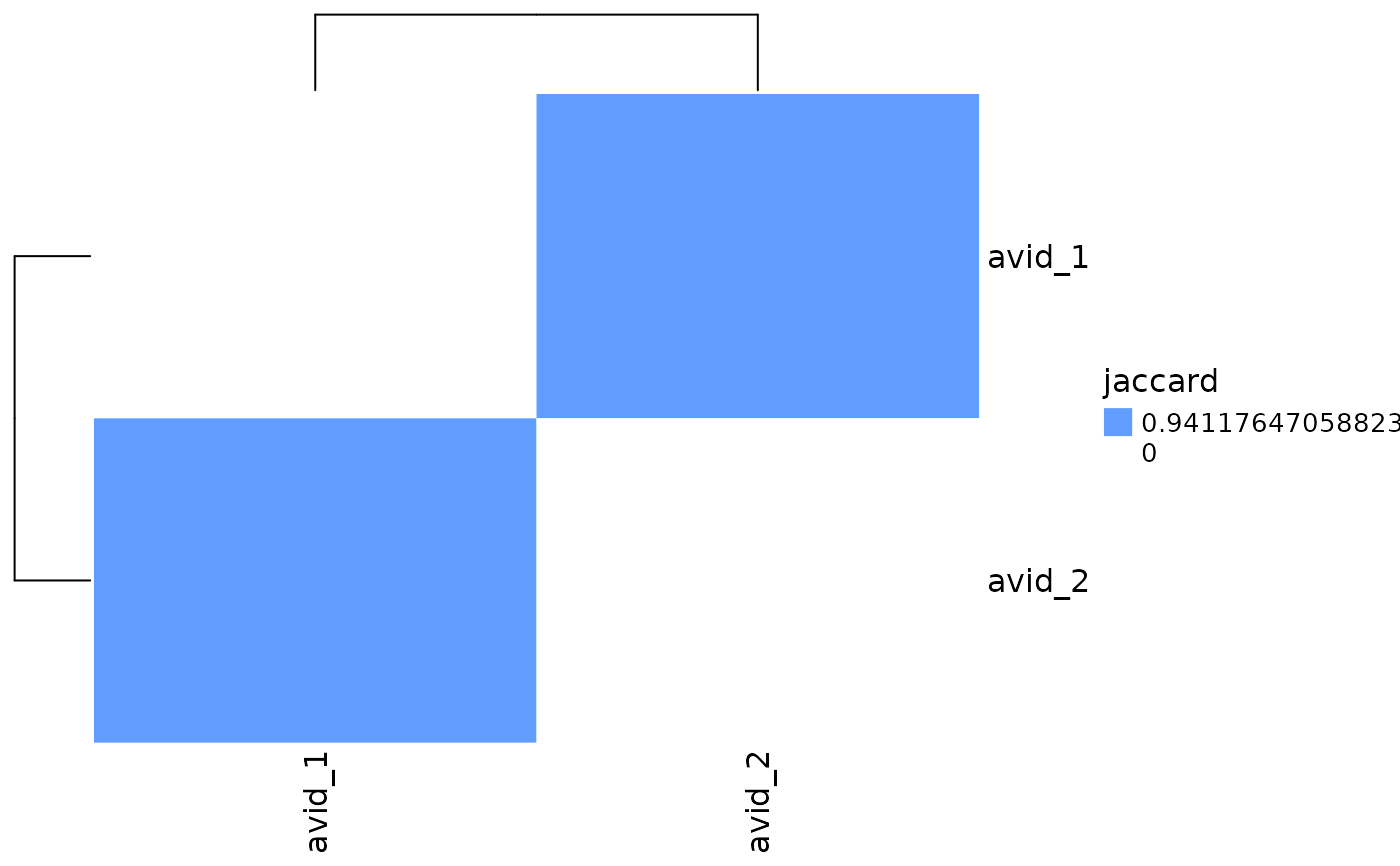
# Plot repertoire overlap
# use clonotype IDs present in 'clonotype_id' column for calculations
plot_similarity(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "orig.ident"
)
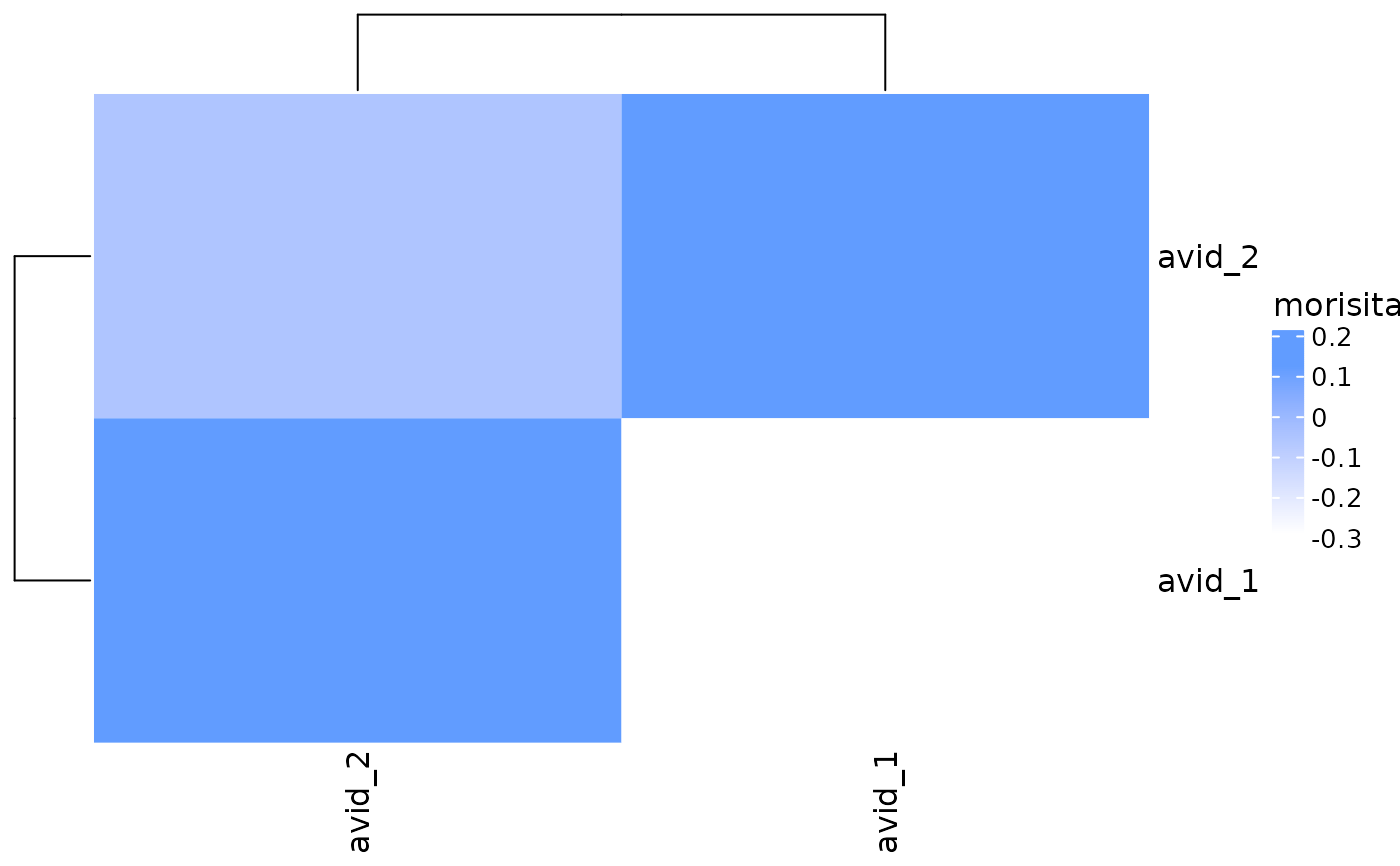
 # Specify method to use for calculating repertoire overlap
plot_similarity(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "orig.ident",
method = abdiv::morisita
)
# Specify method to use for calculating repertoire overlap
plot_similarity(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "orig.ident",
method = abdiv::morisita
)
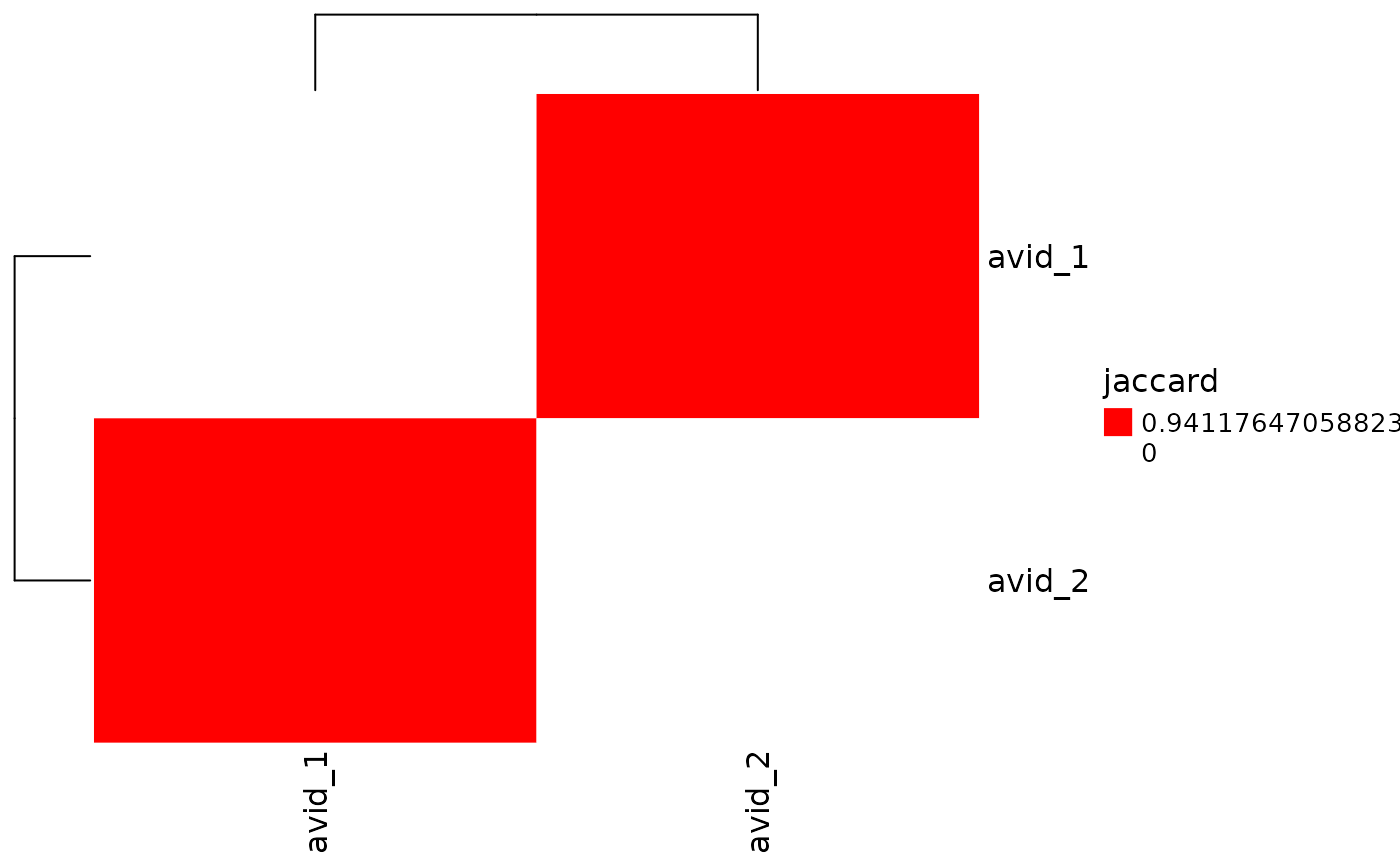
 # Specify colors to use for heatmap
plot_similarity(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "orig.ident",
plot_color = c("white", "red")
)
# Specify colors to use for heatmap
plot_similarity(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "orig.ident",
plot_color = c("white", "red")
)
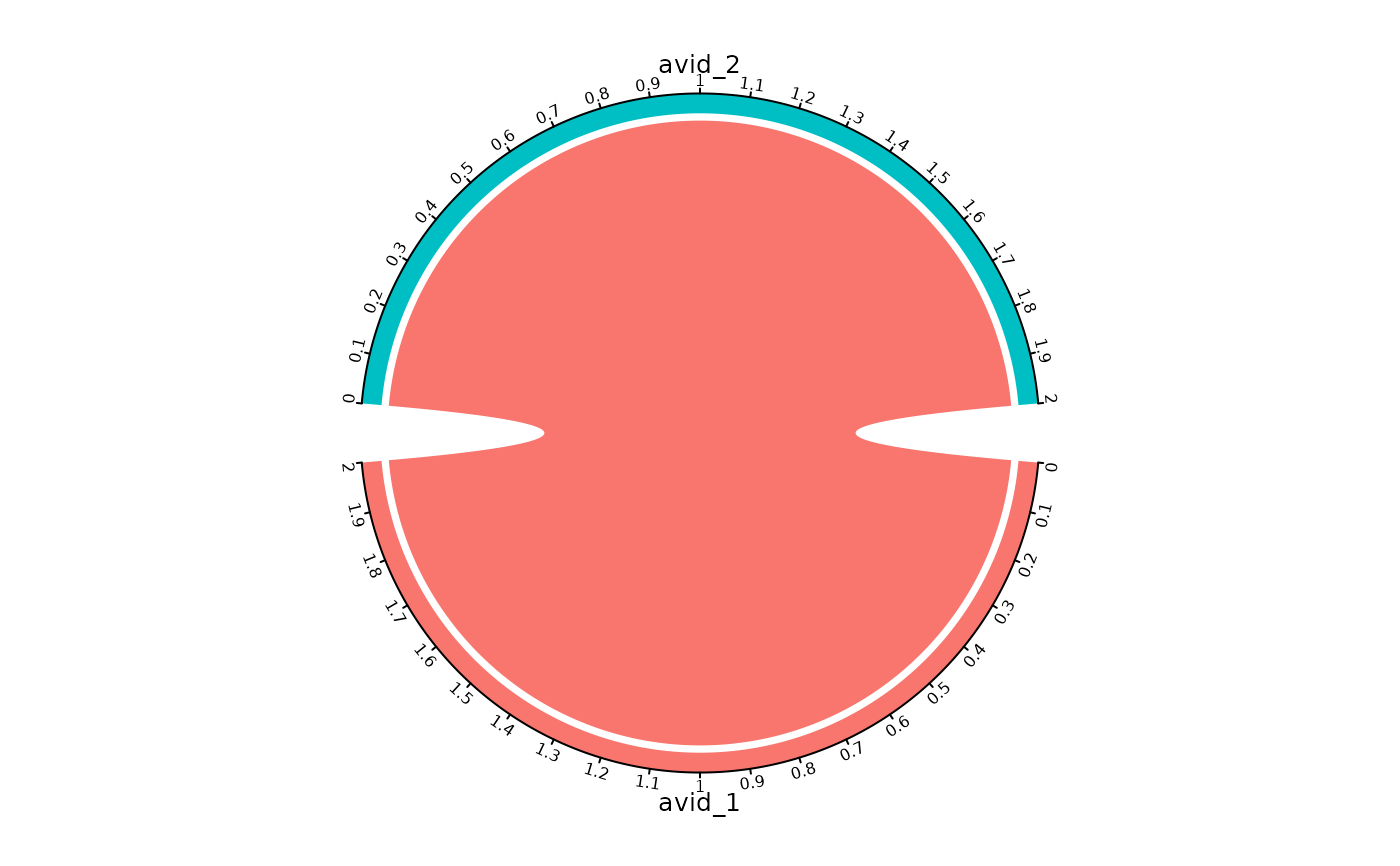
 # Create circos plot
plot_similarity(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "orig.ident",
method = "circos"
)
# Create circos plot
plot_similarity(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "orig.ident",
method = "circos"
)