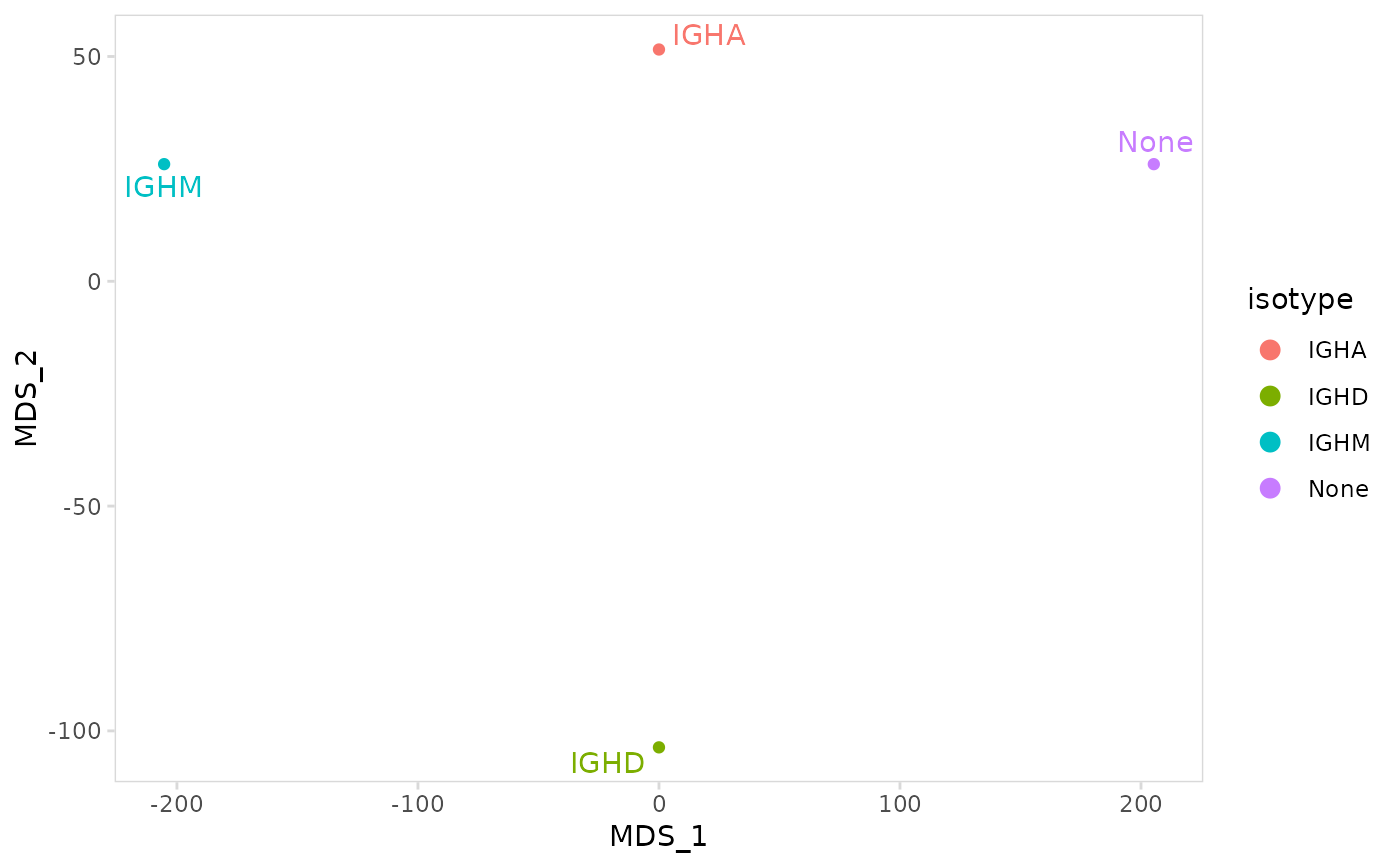
Calculate MDS coordinates based on a beta diversity metric and plot results.
Arguments
- input
Single cell object or data.frame containing V(D)J data. If a data.frame is provided, the cell barcodes should be stored as row names.
- data_col
meta.data column containing values to use for calculating pairwise similarity between clusters, e.g. 'clonotype_id'
- cluster_col
meta.data column containing cluster IDs to use for calculating overlap
- method
Method to use for comparing clusters and calculating MDS coordinates, available methods include:
'jaccard', Jaccard dissimilarity index implemented with
abdiv::jaccard()'horn_morisita', Horn-Morisita index implemented with
abdiv::horn_morisita()
- chain
Chain to use for comparing clusters. To perform calculations using a single chain, the column passed to
data_colmust contain per-chain data such as CDR3 sequences. Set toNULLto include all chains.- chain_col
meta.data column containing chains for each cell
- plot_colors
Character vector containing colors for plotting
- plot_lvls
Levels to use for ordering clusters
- label_points
Label points on plot
- n_label
Location on plot where n label should be added, this can be any combination of the following:
'corner', display the total number of points plotted in the top right corner, the position of the label can be modified by passing
xandyspecifications with thelabel_paramsargument'none', do not display the number of points plotted
- label_params
Named list providing additional parameters to modify n label aesthetics, e.g. list(size = 4, color = "red")
- sep
Separator used for storing per-chain V(D)J data for each cell
- ...
Additional arguments to pass to
ggplot2::geom_point()
Examples
# Calculate MDS coordinates
plot_mds(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "isotype"
)
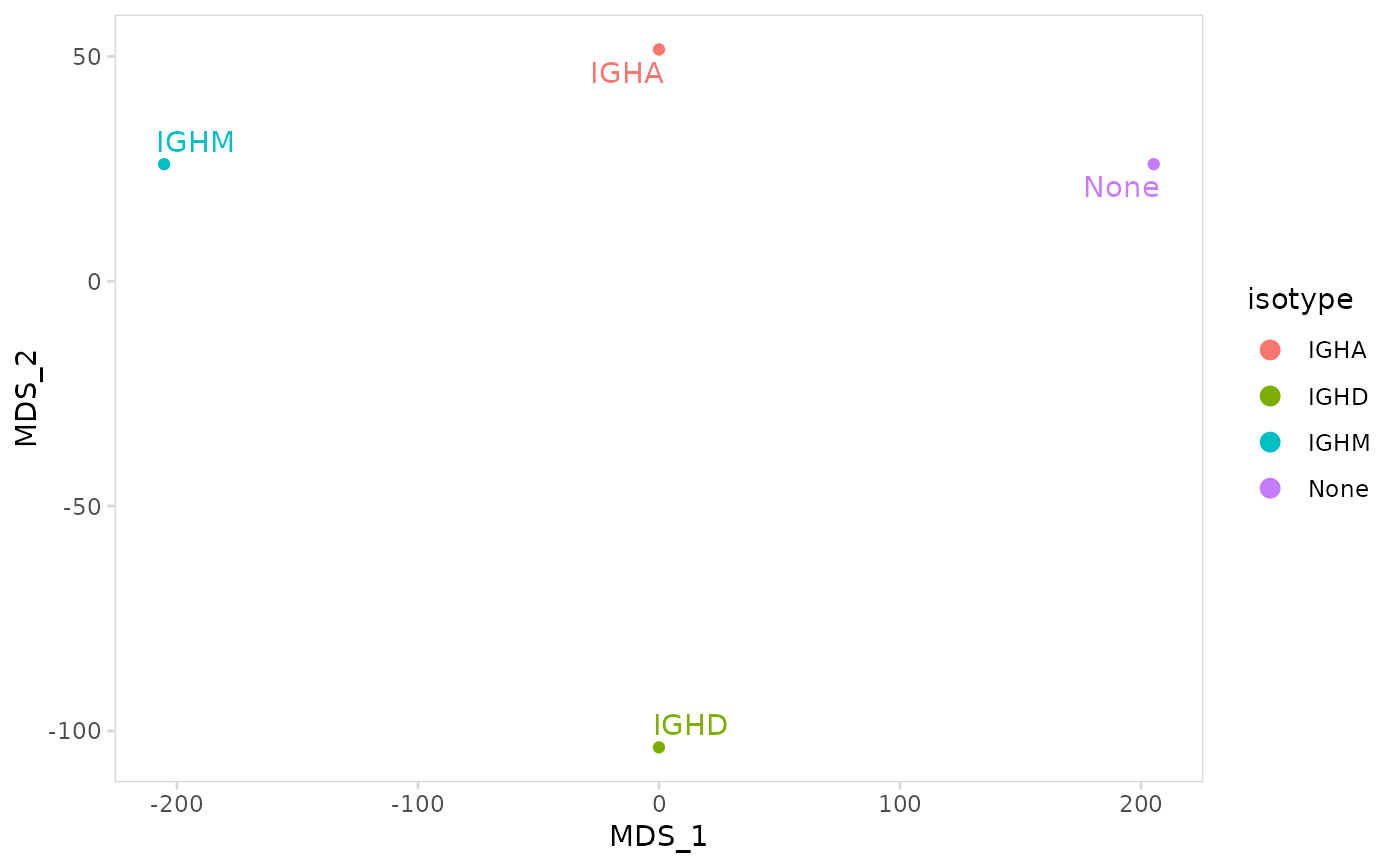 # Calculate MDS coordinates based on IGK CDR3 sequences
plot_mds(
vdj_sce,
data_col = "cdr3",
cluster_col = "isotype",
chain = "IGK"
)
# Calculate MDS coordinates based on IGK CDR3 sequences
plot_mds(
vdj_sce,
data_col = "cdr3",
cluster_col = "isotype",
chain = "IGK"
)
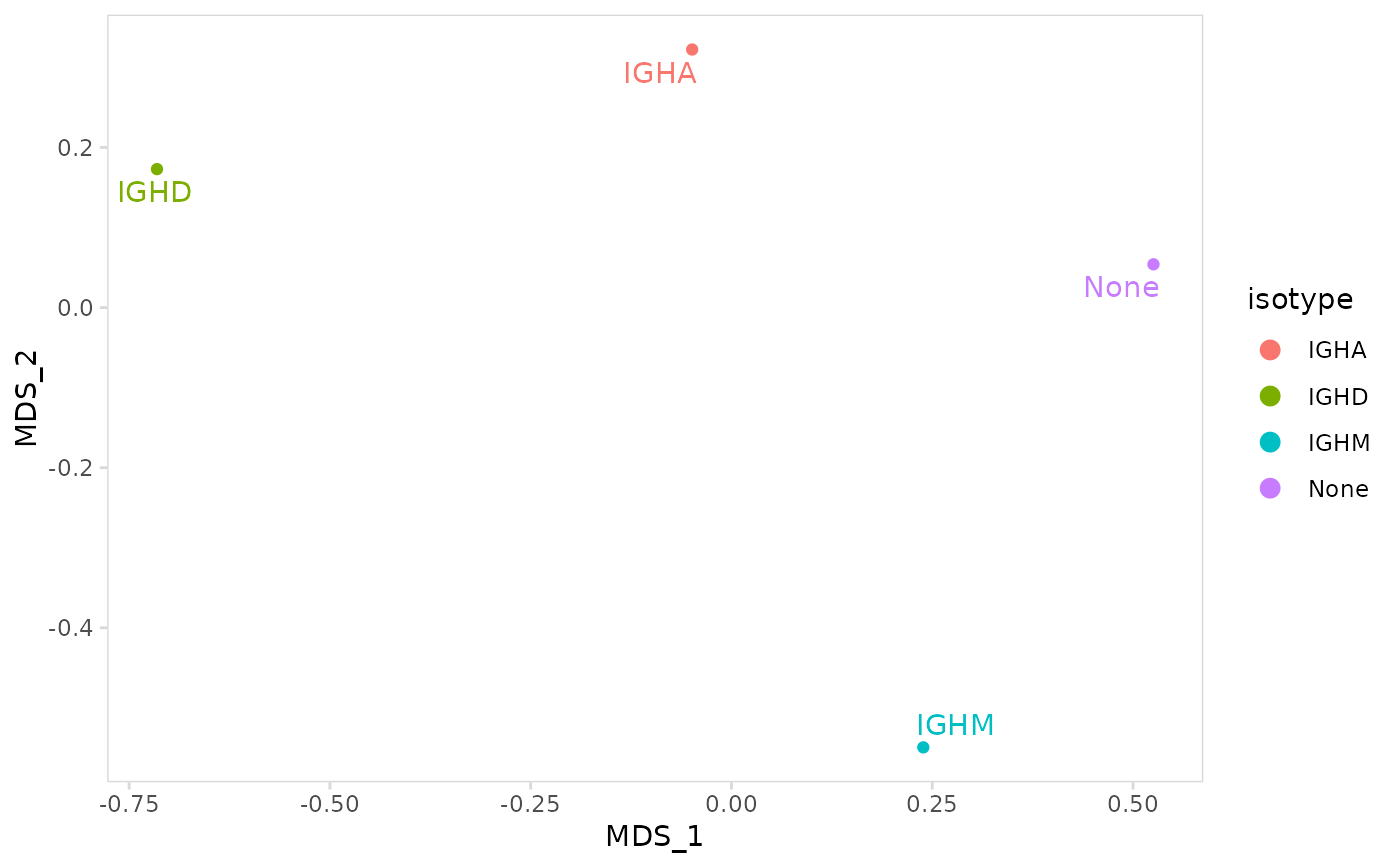 # Calculate repertoire similarity using the Horn-Morisita index
plot_mds(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "isotype",
method = "horn_morisita"
)
# Calculate repertoire similarity using the Horn-Morisita index
plot_mds(
vdj_sce,
data_col = "clonotype_id",
cluster_col = "isotype",
method = "horn_morisita"
)