plot GSEA pathway scores as heatmap, returns a list containing results and plot.
Source:R/utils.R
plot_pathway_gsea.Rdplot GSEA pathway scores as heatmap, returns a list containing results and plot.
Usage
plot_pathway_gsea(
mat,
pathway_list,
n_perm = 1000,
scale = TRUE,
topn = 5,
returning = "both"
)Arguments
- mat
expression matrix
- pathway_list
a list of vectors, each named for a specific pathway, or dataframe
- n_perm
Number of permutation for fgsea function. Defaults to 1000.
- scale
convert expr_mat into zscores prior to running GSEA?, default = TRUE
- topn
number of top pathways to plot
- returning
to return "both" list and plot, or either one
Value
list of matrix and plot, or just plot, matrix of GSEA NES values, cell types as row names, pathways as column names
Examples
gl <- list(
"n" = c("PPBP", "LYZ", "S100A9"),
"a" = c("IGLL5", "GNLY", "FTL")
)
pbmc_avg <- average_clusters(
mat = pbmc_matrix_small,
metadata = pbmc_meta,
cluster_col = "classified"
)
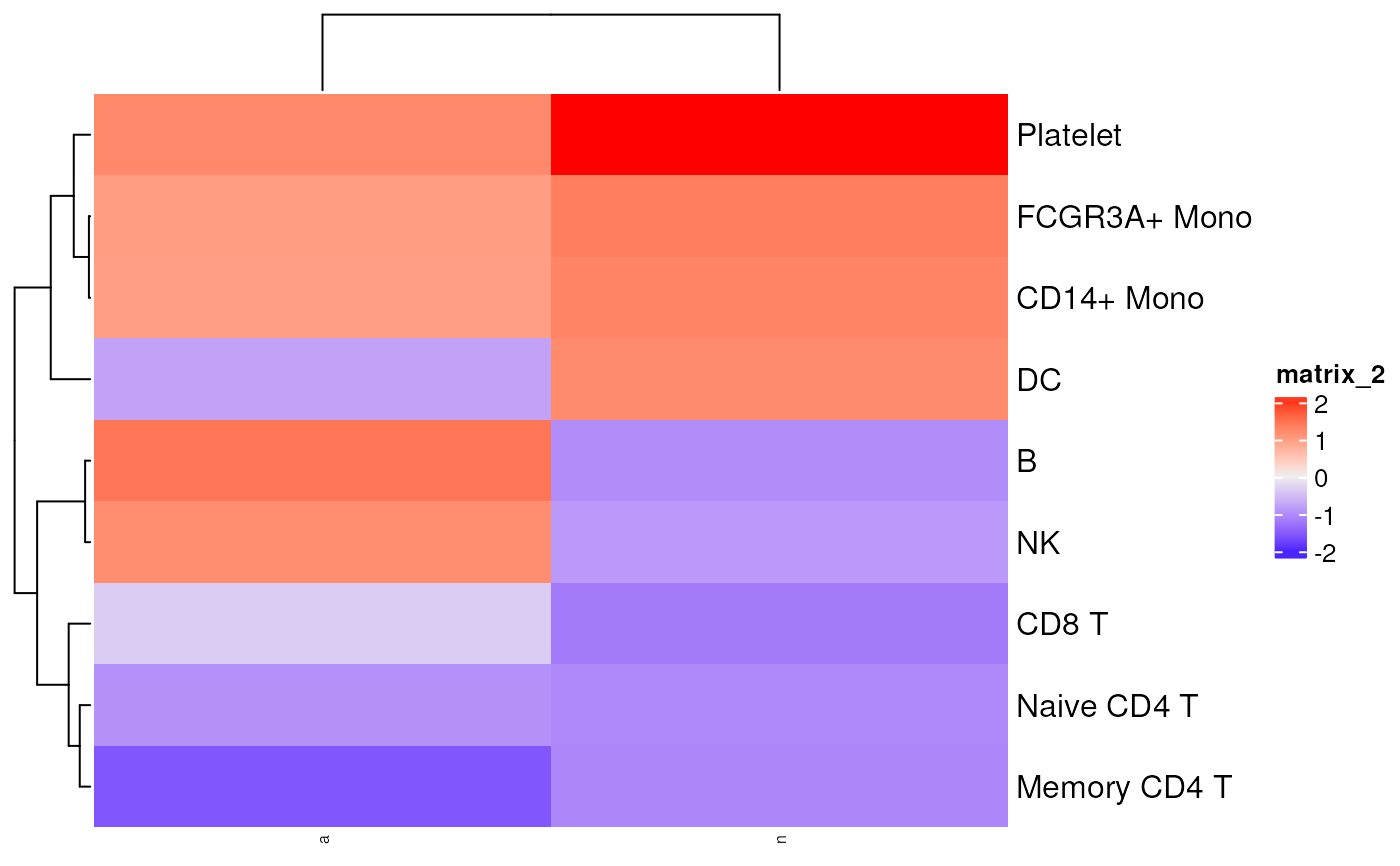
plot_pathway_gsea(
pbmc_avg,
gl,
5
)
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#>
|
| | 0%
|
|======================================================================| 100%
#>
#> [[1]]
#> n a
#> Naive CD4 T -1.0538172 -0.9718567
#> Memory CD4 T -1.0638297 -1.5689151
#> CD14+ Mono 1.2992298 0.9991021
#> B -1.0052358 1.4618113
#> CD8 T -1.2027266 -0.3480414
#> FCGR3A+ Mono 1.3690474 1.0219725
#> NK -0.8783069 1.2010654
#> DC 1.2336765 -0.7802910
#> Platelet 2.2022008 1.2544830
#>
#> [[2]]
 #>
#>