Identify intervals within a specified distance.
Details
input tbls are grouped by chrom by default, and additional
groups can be added using dplyr::group_by(). For example,
grouping by strand will constrain analyses to the same strand. To
compare opposing strands across two tbls, strands on the y tbl can
first be inverted using flip_strands().
See also
https://bedtools.readthedocs.io/en/latest/content/tools/window.html
Other multiple set operations:
bed_closest(),
bed_coverage(),
bed_intersect(),
bed_map(),
bed_subtract()
Examples
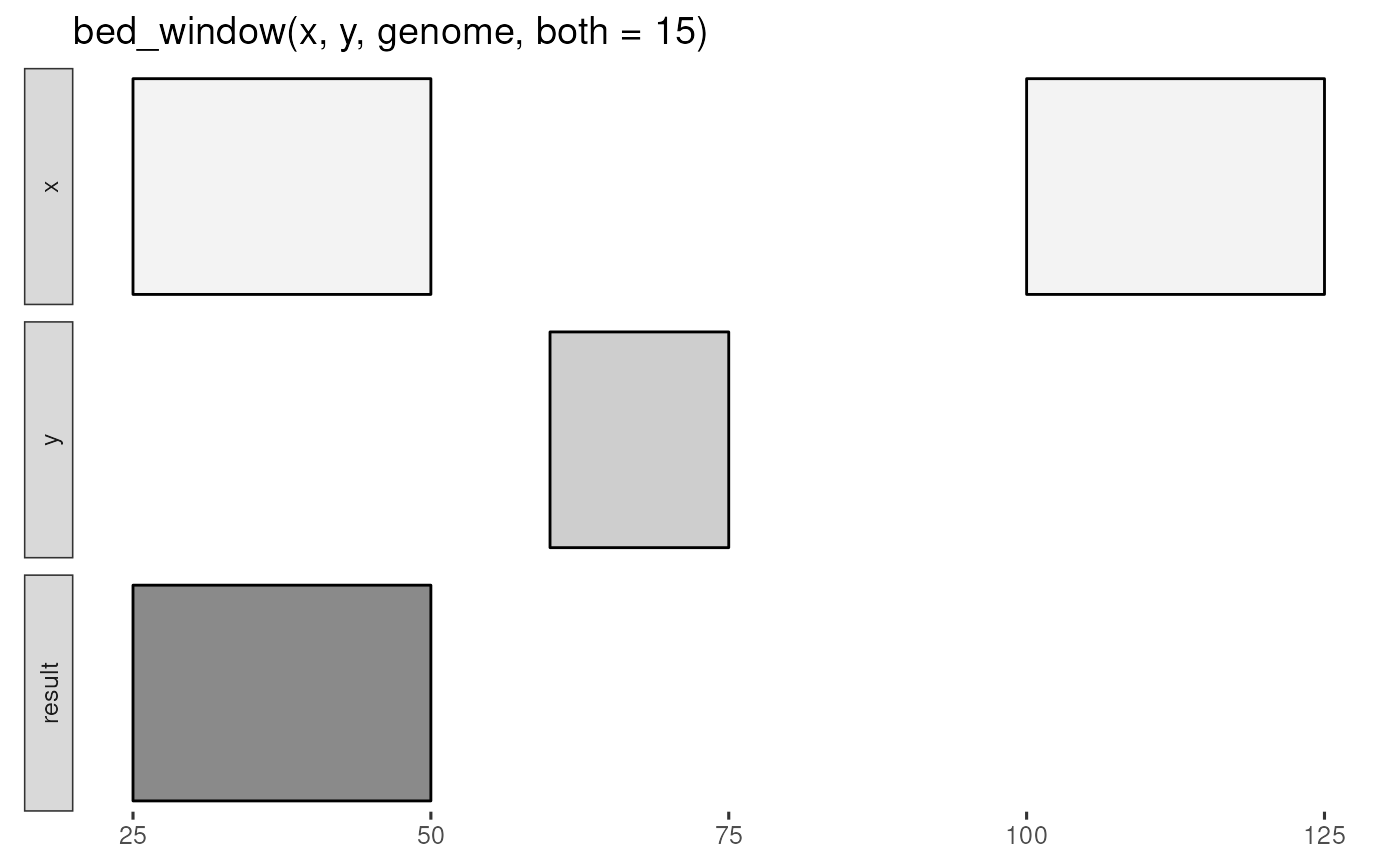
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 25, 50,
"chr1", 100, 125
)
y <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 60, 75
)
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 125
)
bed_glyph(bed_window(x, y, genome, both = 15))
 x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 10, 100,
"chr2", 200, 400,
"chr2", 300, 500,
"chr2", 800, 900
)
y <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 150, 400,
"chr2", 230, 430,
"chr2", 350, 430
)
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 500,
"chr2", 1000
)
bed_window(x, y, genome, both = 100)
#> # A tibble: 4 × 7
#> chrom start.x end.x start.y end.y .source .overlap
#> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <int>
#> 1 chr2 200 400 230 430 1 200
#> 2 chr2 200 400 350 430 1 80
#> 3 chr2 300 500 230 430 1 200
#> 4 chr2 300 500 350 430 1 80
# add a `.dist` column to the output
if (FALSE) { # \dontrun{
bed_window(x, y, genome, both = 200) |>
mutate(
.dist = case_when(
.overlap == 0 ~ abs(pmax(start.x, start.y) - pmin(end.x, end.y)),
.default = 0
)
)
} # }
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 10, 100,
"chr2", 200, 400,
"chr2", 300, 500,
"chr2", 800, 900
)
y <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 150, 400,
"chr2", 230, 430,
"chr2", 350, 430
)
genome <- tibble::tribble(
~chrom, ~size,
"chr1", 500,
"chr2", 1000
)
bed_window(x, y, genome, both = 100)
#> # A tibble: 4 × 7
#> chrom start.x end.x start.y end.y .source .overlap
#> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <int>
#> 1 chr2 200 400 230 430 1 200
#> 2 chr2 200 400 350 430 1 80
#> 3 chr2 300 500 230 430 1 200
#> 4 chr2 300 500 350 430 1 80
# add a `.dist` column to the output
if (FALSE) { # \dontrun{
bed_window(x, y, genome, both = 200) |>
mutate(
.dist = case_when(
.overlap == 0 ~ abs(pmax(start.x, start.y) - pmin(end.x, end.y)),
.default = 0
)
)
} # }
