Identify closest intervals.
Usage
bed_closest(x, y, overlap = TRUE, suffix = c(".x", ".y"))Value
ivl_df with additional columns:
.overlapamount of overlap with overlapping interval. Non-overlapping or adjacent intervals have an overlap of 0..overlapwill not be included in the output ifoverlap = FALSE..distdistance to closest interval. Negative distances denote upstream intervals. Book-ended intervals have a distance of 1.
Details
input tbls are grouped by chrom by default, and additional
groups can be added using dplyr::group_by(). For example,
grouping by strand will constrain analyses to the same strand. To
compare opposing strands across two tbls, strands on the y tbl can
first be inverted using flip_strands().
Note
For each interval in x bed_closest() returns overlapping intervals from y
and the closest non-intersecting y interval. Setting overlap = FALSE will
report the closest non-intersecting y intervals, ignoring any overlapping y
intervals.
See also
https://bedtools.readthedocs.io/en/latest/content/tools/closest.html
Other multiple set operations:
bed_coverage(),
bed_intersect(),
bed_map(),
bed_subtract(),
bed_window()
Examples
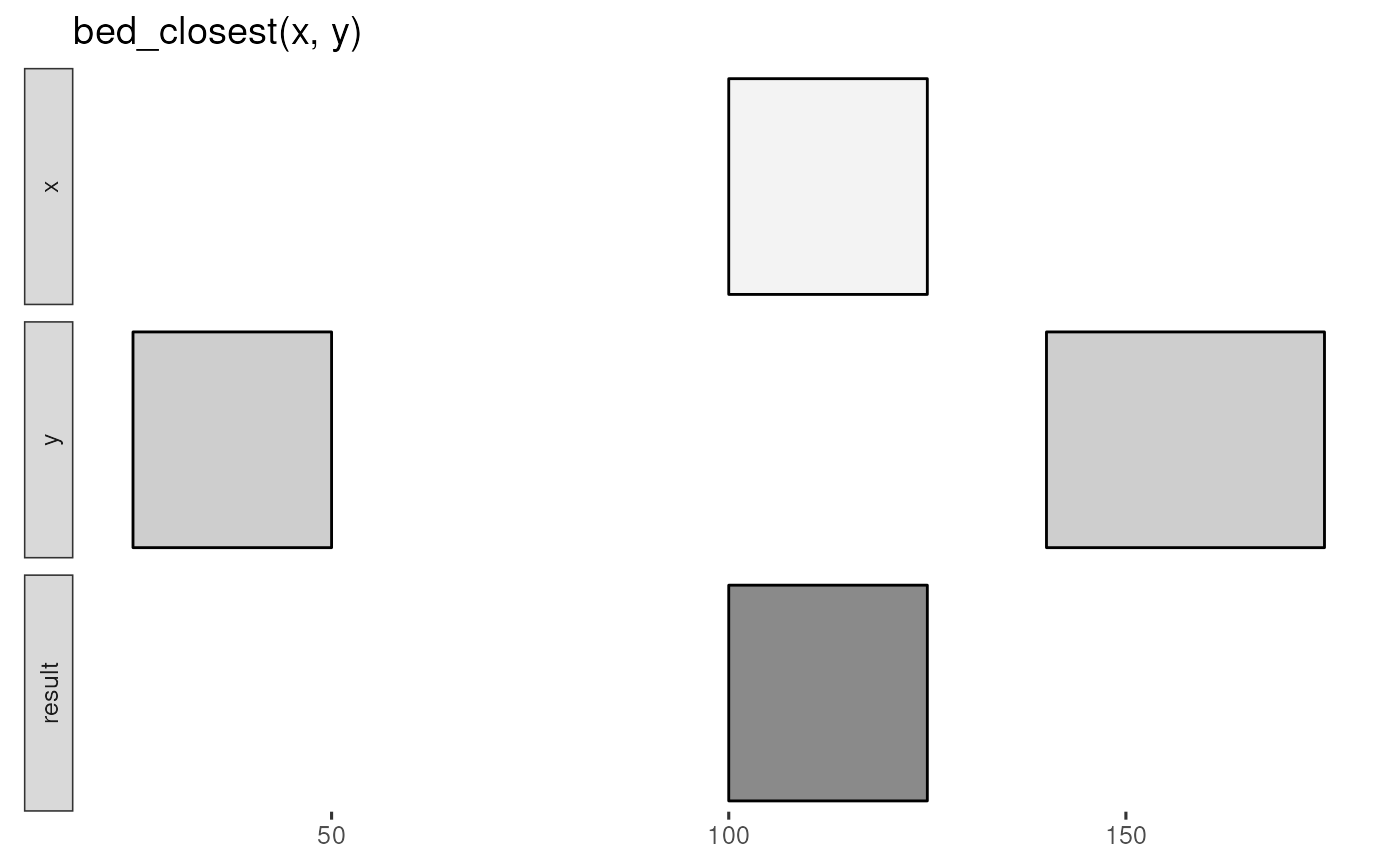
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 100, 125
)
y <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 25, 50,
"chr1", 140, 175
)
bed_glyph(bed_closest(x, y))
 x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 500, 600,
"chr2", 5000, 6000
)
y <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 100, 200,
"chr1", 150, 200,
"chr1", 550, 580,
"chr2", 7000, 8500
)
bed_closest(x, y)
#> # A tibble: 4 × 7
#> chrom start.x end.x start.y end.y .overlap .dist
#> <chr> <dbl> <dbl> <dbl> <dbl> <int> <int>
#> 1 chr1 500 600 550 580 30 0
#> 2 chr1 500 600 100 200 0 -301
#> 3 chr1 500 600 150 200 0 -301
#> 4 chr2 5000 6000 7000 8500 0 1001
bed_closest(x, y, overlap = FALSE)
#> # A tibble: 3 × 6
#> chrom start.x end.x start.y end.y .dist
#> <chr> <dbl> <dbl> <dbl> <dbl> <int>
#> 1 chr1 500 600 100 200 -301
#> 2 chr1 500 600 150 200 -301
#> 3 chr2 5000 6000 7000 8500 1001
# Report distance based on strand
x <- tibble::tribble(
~chrom, ~start, ~end, ~name, ~score, ~strand,
"chr1", 10, 20, "a", 1, "-"
)
y <- tibble::tribble(
~chrom, ~start, ~end, ~name, ~score, ~strand,
"chr1", 8, 9, "b", 1, "+",
"chr1", 21, 22, "b", 1, "-"
)
res <- bed_closest(x, y)
# convert distance based on strand
res$.dist_strand <- ifelse(res$strand.x == "+", res$.dist, -(res$.dist))
res
#> # A tibble: 2 × 14
#> chrom start.x end.x name.x score.x strand.x start.y end.y name.y score.y
#> <chr> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 chr1 10 20 a 1 - 21 22 b 1
#> 2 chr1 10 20 a 1 - 8 9 b 1
#> # ℹ 4 more variables: strand.y <chr>, .overlap <int>, .dist <int>,
#> # .dist_strand <int>
# report absolute distances
res$.abs_dist <- abs(res$.dist)
res
#> # A tibble: 2 × 15
#> chrom start.x end.x name.x score.x strand.x start.y end.y name.y score.y
#> <chr> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 chr1 10 20 a 1 - 21 22 b 1
#> 2 chr1 10 20 a 1 - 8 9 b 1
#> # ℹ 5 more variables: strand.y <chr>, .overlap <int>, .dist <int>,
#> # .dist_strand <int>, .abs_dist <int>
x <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 500, 600,
"chr2", 5000, 6000
)
y <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 100, 200,
"chr1", 150, 200,
"chr1", 550, 580,
"chr2", 7000, 8500
)
bed_closest(x, y)
#> # A tibble: 4 × 7
#> chrom start.x end.x start.y end.y .overlap .dist
#> <chr> <dbl> <dbl> <dbl> <dbl> <int> <int>
#> 1 chr1 500 600 550 580 30 0
#> 2 chr1 500 600 100 200 0 -301
#> 3 chr1 500 600 150 200 0 -301
#> 4 chr2 5000 6000 7000 8500 0 1001
bed_closest(x, y, overlap = FALSE)
#> # A tibble: 3 × 6
#> chrom start.x end.x start.y end.y .dist
#> <chr> <dbl> <dbl> <dbl> <dbl> <int>
#> 1 chr1 500 600 100 200 -301
#> 2 chr1 500 600 150 200 -301
#> 3 chr2 5000 6000 7000 8500 1001
# Report distance based on strand
x <- tibble::tribble(
~chrom, ~start, ~end, ~name, ~score, ~strand,
"chr1", 10, 20, "a", 1, "-"
)
y <- tibble::tribble(
~chrom, ~start, ~end, ~name, ~score, ~strand,
"chr1", 8, 9, "b", 1, "+",
"chr1", 21, 22, "b", 1, "-"
)
res <- bed_closest(x, y)
# convert distance based on strand
res$.dist_strand <- ifelse(res$strand.x == "+", res$.dist, -(res$.dist))
res
#> # A tibble: 2 × 14
#> chrom start.x end.x name.x score.x strand.x start.y end.y name.y score.y
#> <chr> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 chr1 10 20 a 1 - 21 22 b 1
#> 2 chr1 10 20 a 1 - 8 9 b 1
#> # ℹ 4 more variables: strand.y <chr>, .overlap <int>, .dist <int>,
#> # .dist_strand <int>
# report absolute distances
res$.abs_dist <- abs(res$.dist)
res
#> # A tibble: 2 × 15
#> chrom start.x end.x name.x score.x strand.x start.y end.y name.y score.y
#> <chr> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 chr1 10 20 a 1 - 21 22 b 1
#> 2 chr1 10 20 a 1 - 8 9 b 1
#> # ℹ 5 more variables: strand.y <chr>, .overlap <int>, .dist <int>,
#> # .dist_strand <int>, .abs_dist <int>
