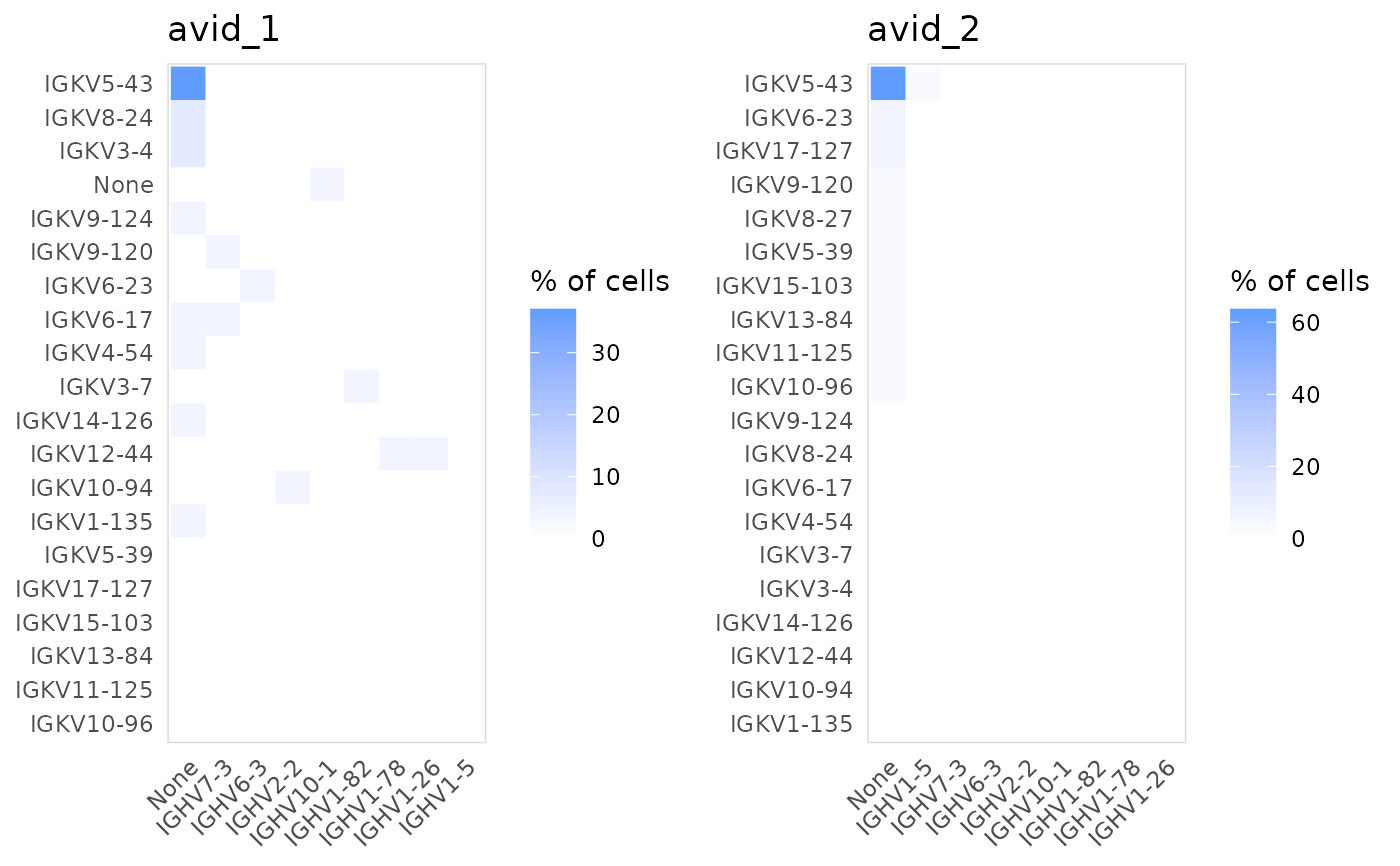
Plot the paired usage of V(D)J segments across two chains. For example, plot_gene_pairs() can be used to plot the frequency that different TRA and TRB V segments appear together.
Usage
plot_gene_pairs(
input,
data_col,
chains,
cluster_col = NULL,
genes = 20,
chain_col = global$chain_col,
method = "heatmap",
units = "percent",
return_list = FALSE,
sep = global$sep,
plot_colors = NULL,
plot_lvls = NULL,
trans = "identity",
rotate_labels = FALSE,
panel_nrow = NULL,
...
)Arguments
- input
Object containing V(D)J data. If a data.frame is provided, the cell barcodes should be stored as row names.
- data_col
meta.data column containing V(D)J genes identified for each clonotype.
- chains
Chains to use for calculating usage of different gene pairs. This should be a character vector containing the two chains to use for calculations, e.g.
c("IGH", "IGK").- cluster_col
meta.data column containing cell clusters to use when calculating gene usage
- genes
An integer specifying the number of genes to plot, or a vector giving the names of genes to include.
- chain_col
meta.data column containing chains for each cell
- method
Method to use for plotting, possible values are:
'heatmap', create a heatmap, this is the default when two columns are passed to the data_cols argument
'circos', create a circos plot, this requires two columns to be provided to the data_cols argument
- units
Units to show on scale, either 'frequency' or 'percent'
- return_list
Should a list of plots be returned, if FALSE plots will be combined and arranged into panels
- sep
Separator used for storing per-chain V(D)J data for each cell
- plot_colors
Character vector containing colors to use for plot. If a bar graph is created this will specify how to color cell clusters. For a heatmap, these colors will be used to generate the color gradient.
- plot_lvls
Levels to use for ordering clusters
- trans
Transformation to use when plotting segment usage, e.g. 'log10'. By default values are not transformed, refer to
ggplot2::continuous_scale()for more options.- rotate_labels
Should labels on circos plot be rotated to reduce overlapping text
- panel_nrow
The number of rows to use for arranging plots when return_list is FALSE
- ...
Additional arguments to pass to plotting function,
ggplot2::geom_tile()for heatmap,circlize::chordDiagram()for circos plot
Examples
# Plot the frequency of different V genes for IGH and IGK chains
plot_gene_pairs(
vdj_sce,
data_col = "v_gene",
chains = c("IGH", "IGK"),
cluster_col = "orig.ident"
)