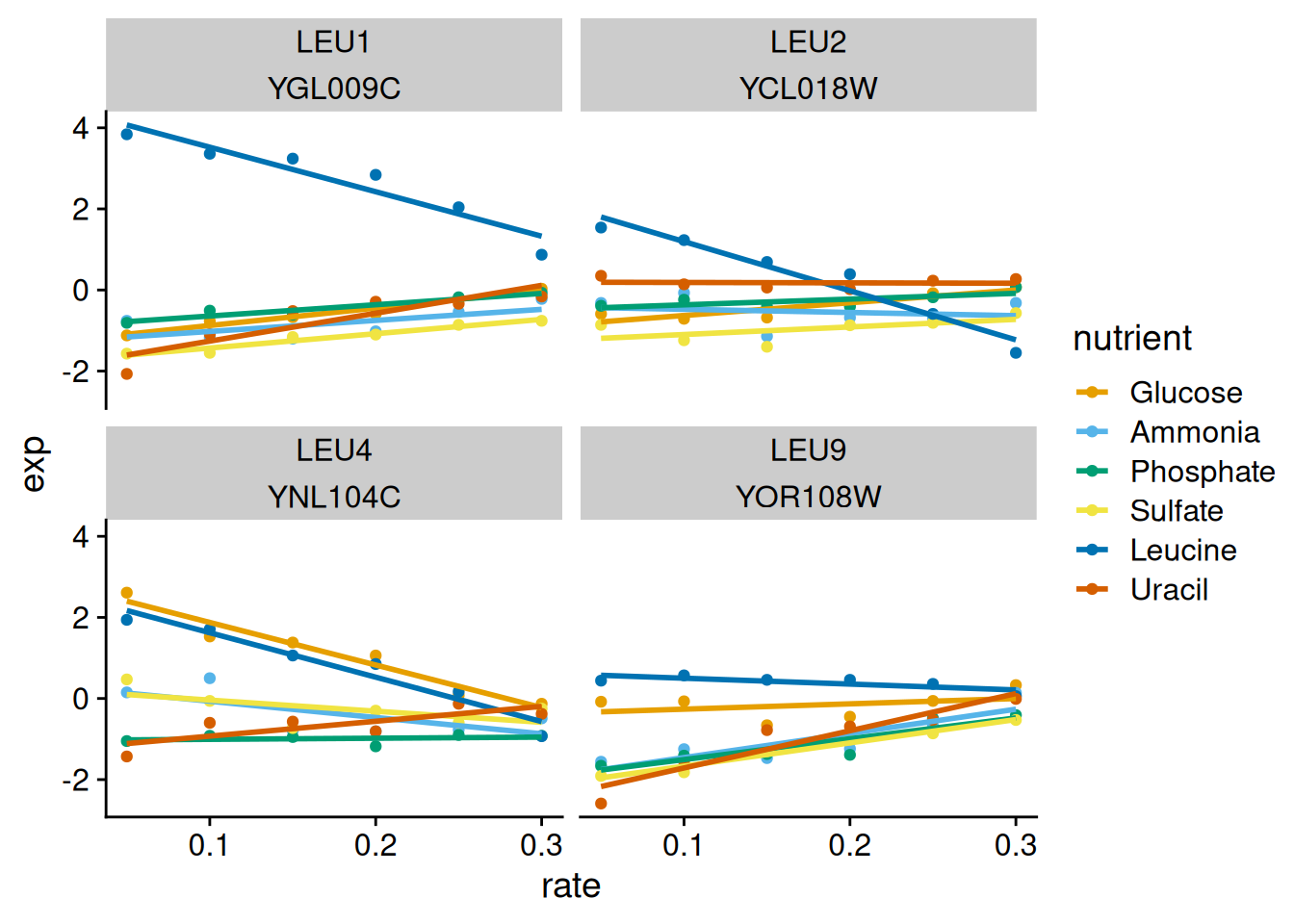
library(yeastgenes)
library(tidyverse)
library(cowplot)
library(ggokabeito)
yeast_gene_exp |>
left_join(yeast_go_terms) |>
filter(go_biological_process == "leucine biosynthesis") |>
ggplot(
aes(
rate,
exp,
color = nutrient
)
) +
geom_point() +
geom_smooth(method = "lm", se = FALSE) +
facet_wrap(~ common_name + systematic_name) +
theme_cowplot() +
scale_color_okabe_ito()