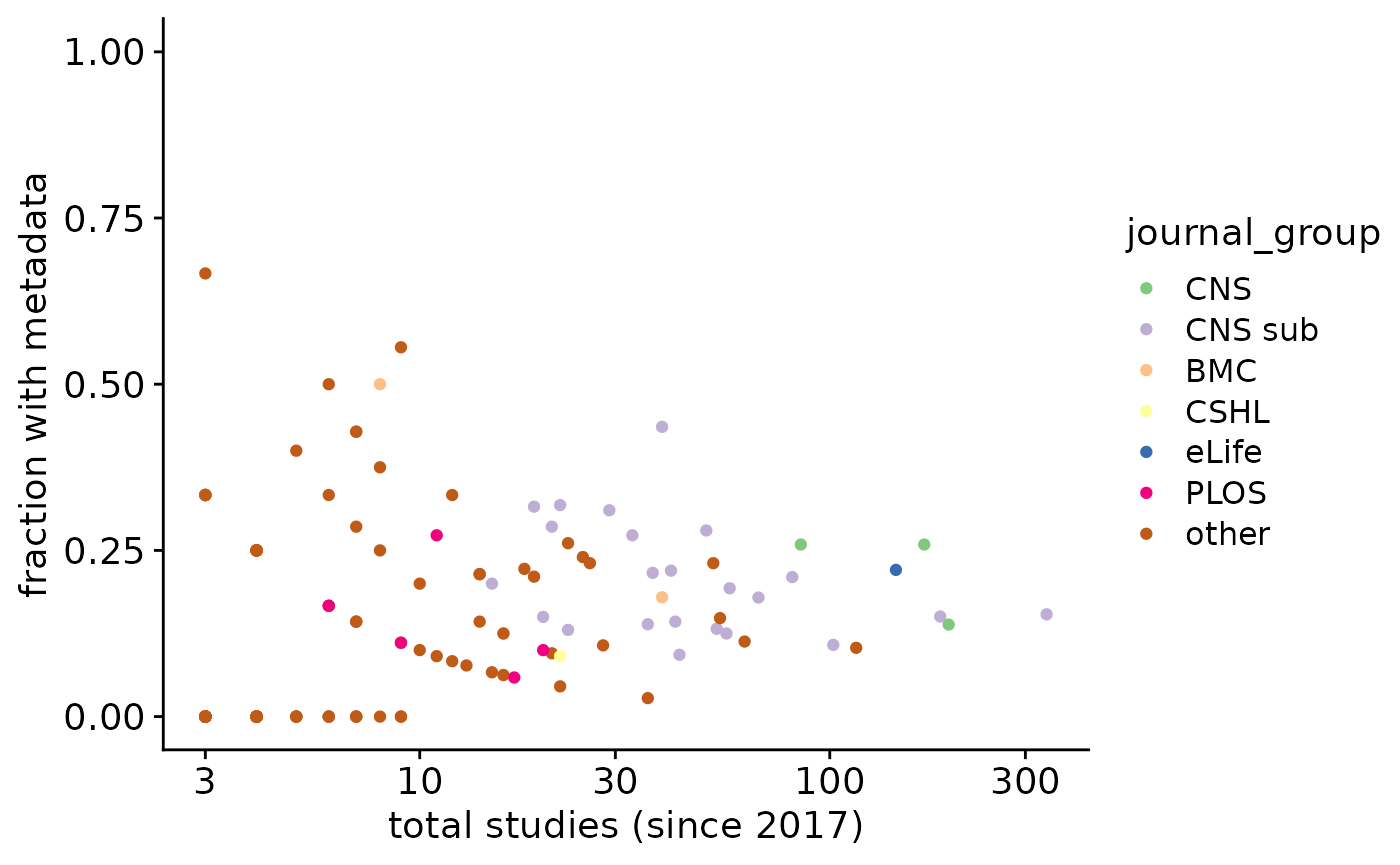Assessment of missing cell-level metadata in single cell GEO records
Rui Fu
RNA Bioscience Initative, University of Colorado School of Medicine2022-02-21
get_geo.RmdCurrent GEO query used:
"expression profiling by high throughput sequencing" AND
("single nuclei" OR "single cell" OR "scRNAseq" OR "scRNA-seq" OR "snRNAseq" OR "snRNA-seq")#> [1] "022122"
#> Response [https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=gds&query_key=1&WebEnv=MCID_6212fe0866927a572d71240b]
#> Date: 2022-02-21 02:50
#> Status: 200
#> Content-Type: text/plain; charset=UTF-8
#> Size: 6.52 MB
#> <ON DISK> /home/runner/work/someta/someta/inst/extdata/022122/gds_result_022122.txtParse GEO query
Total entries: 7805
Number of entries filtered out because key words were not found: 926
Merged super and subseries: 593
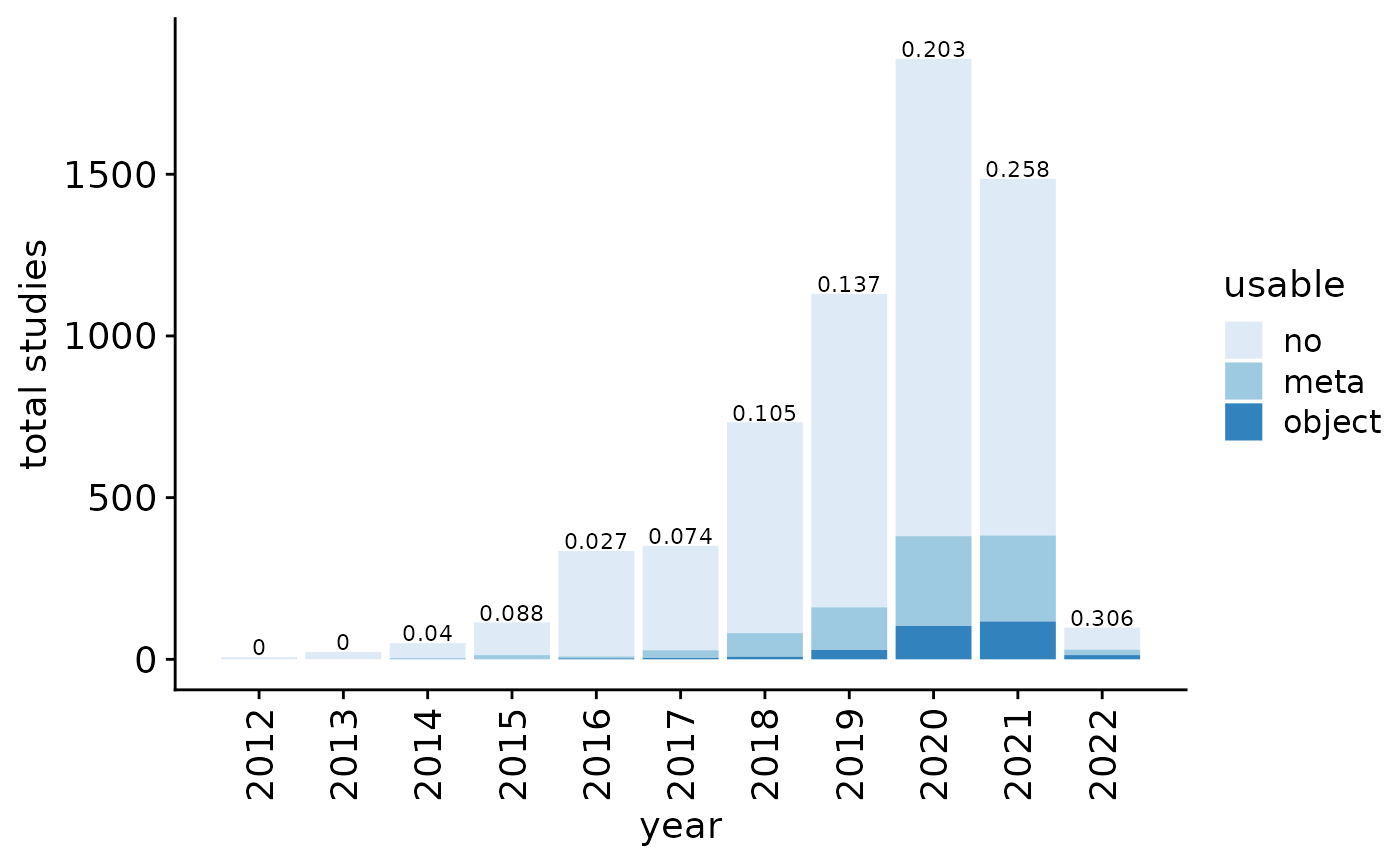
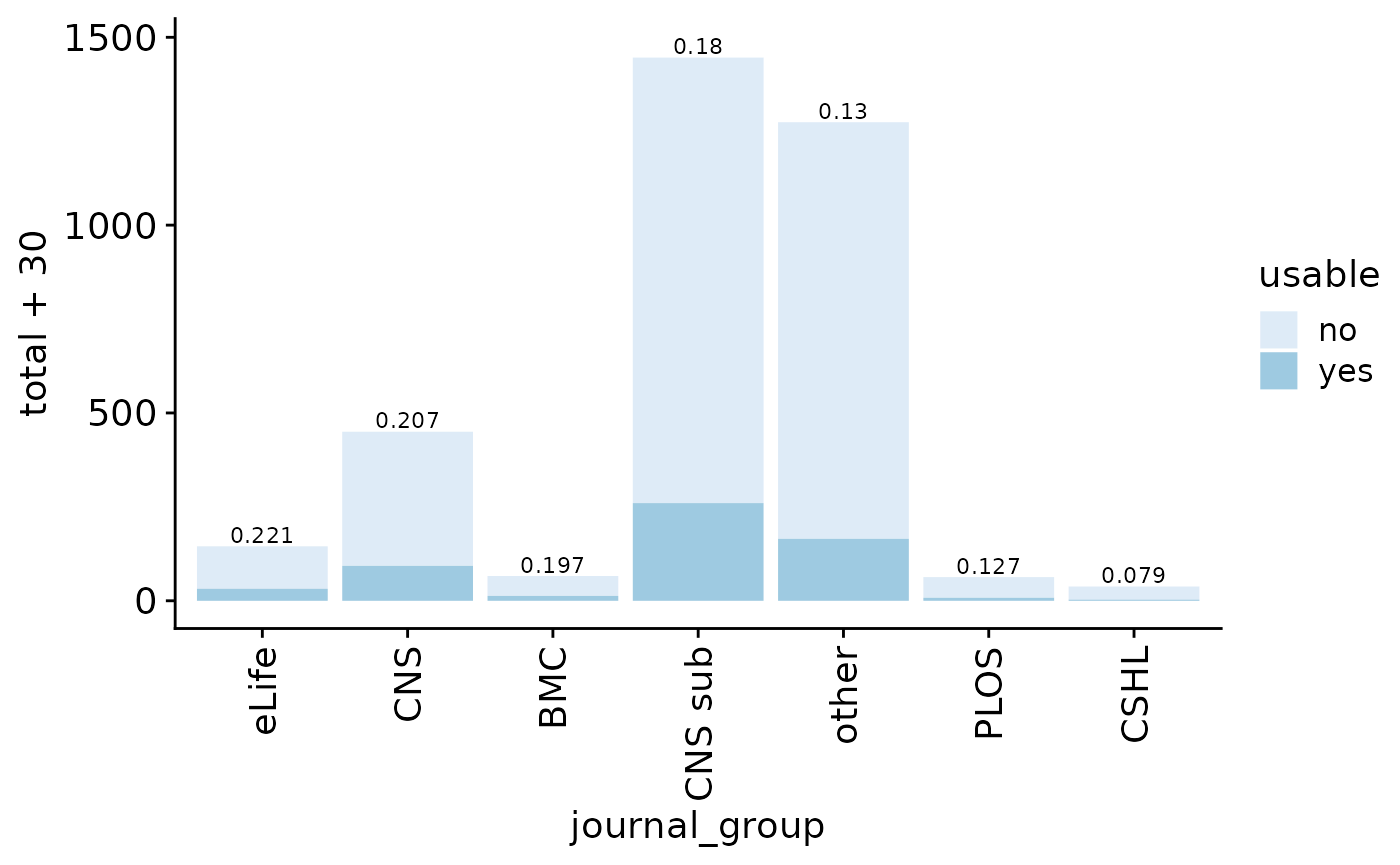
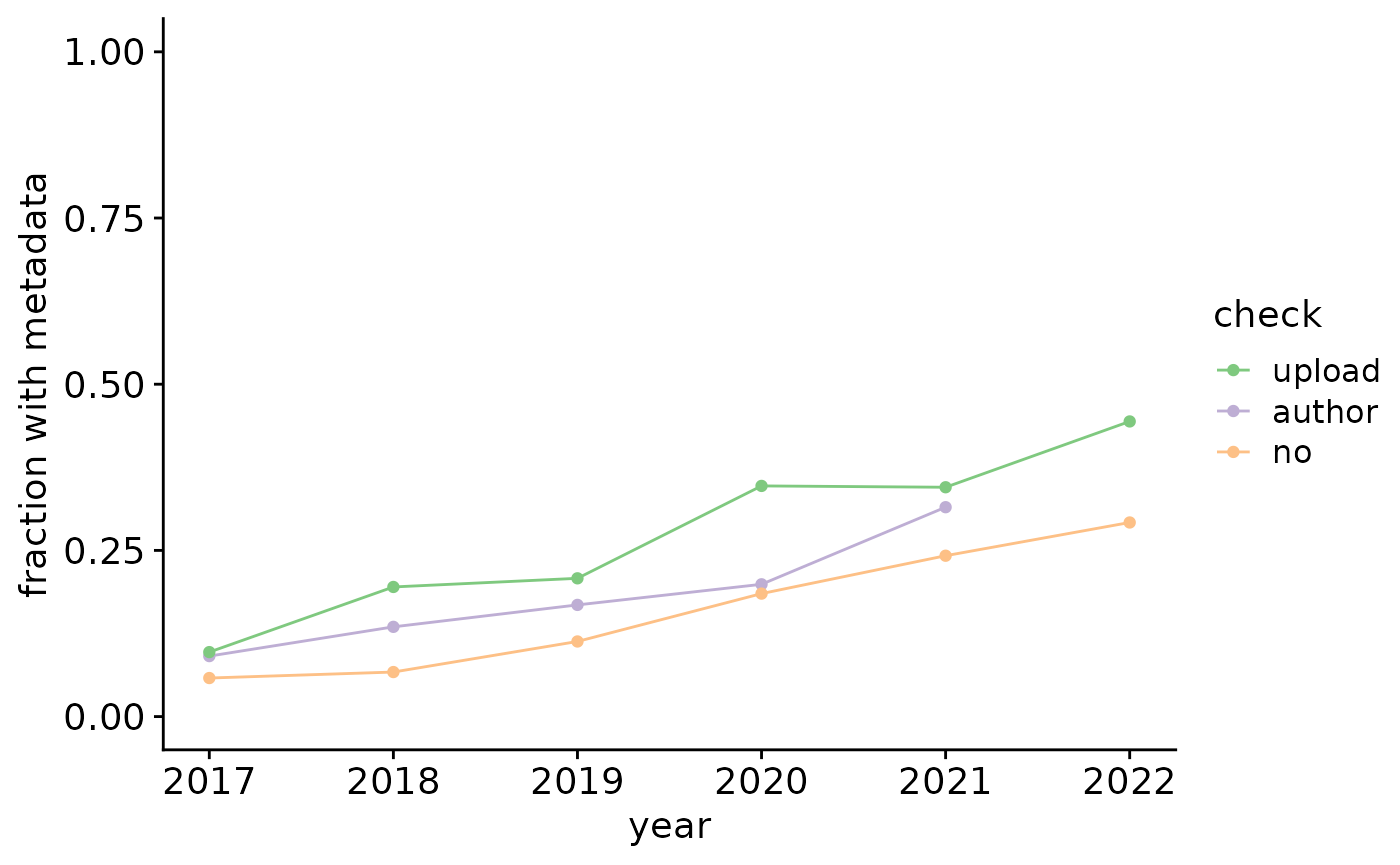
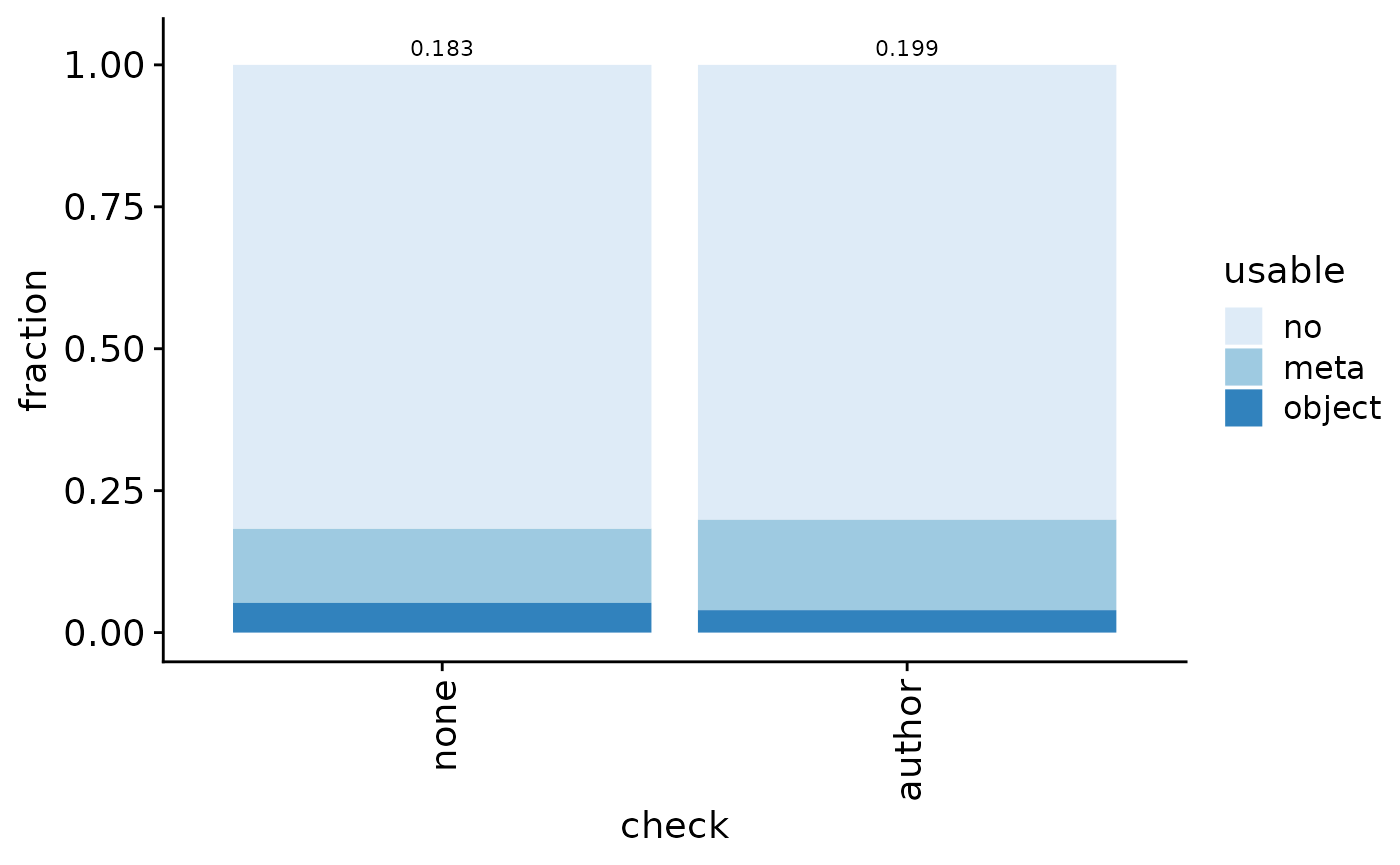
The fraction of GEO entries with potential metadata (file with “meta”, “annot”, “clustering”, “colData”, or “type” in filename or rda/rds/rdata/h5ad/loom files) is 0.1760905. Note however that these terms include some false positives (such as gene annotation file, patient metadata, and phenotype table), which we manually inspected and corrected (false positive fraction at 0.0491803). Final fraction: 0.1726979