possible_annotations <- builtin_annotations()
# grep to keep those containing "hg19"
hg19_annots <- grep("hg19_genes", possible_annotations, value = T)
# WHY DID WE PICK hg19?
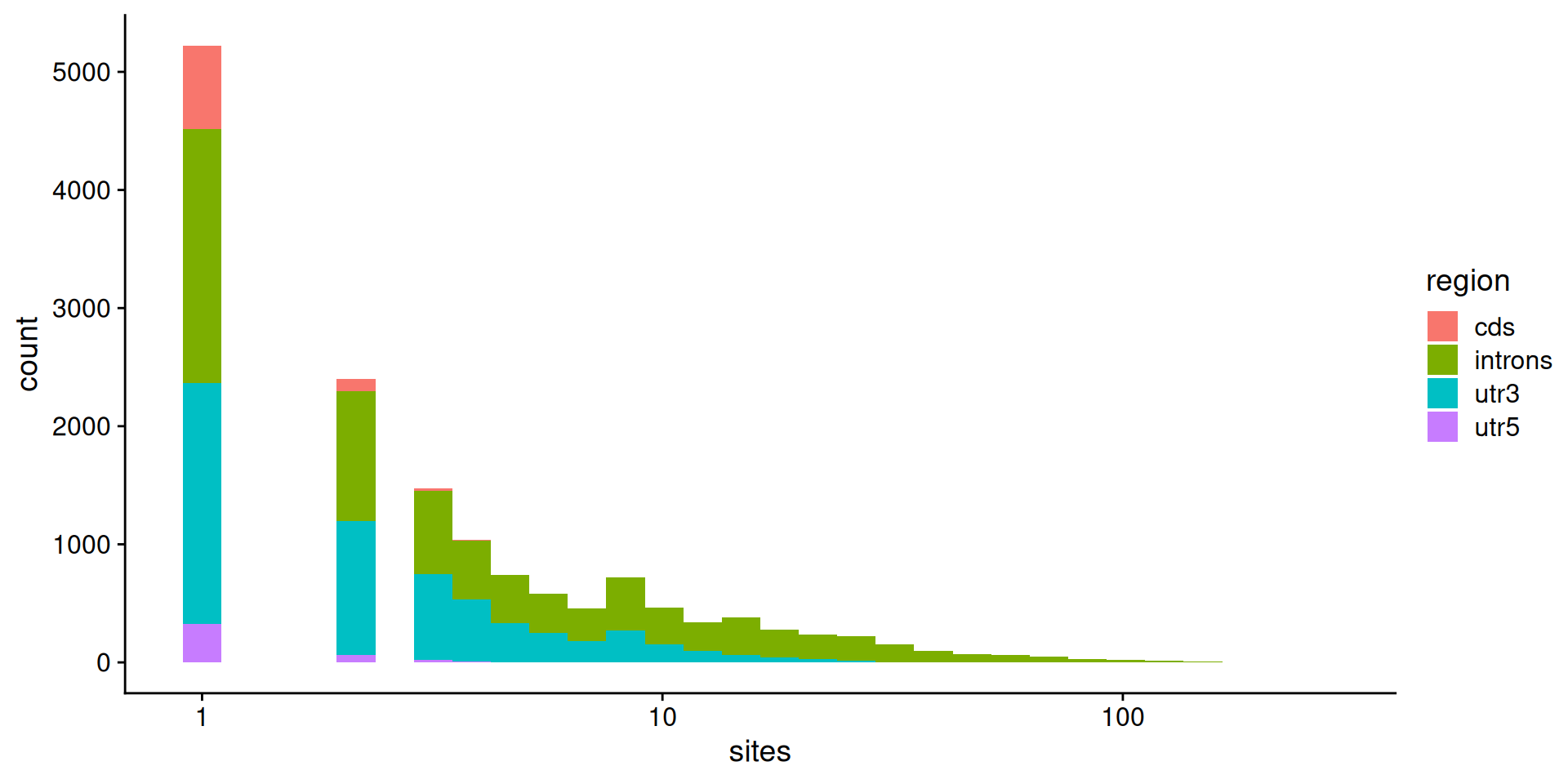
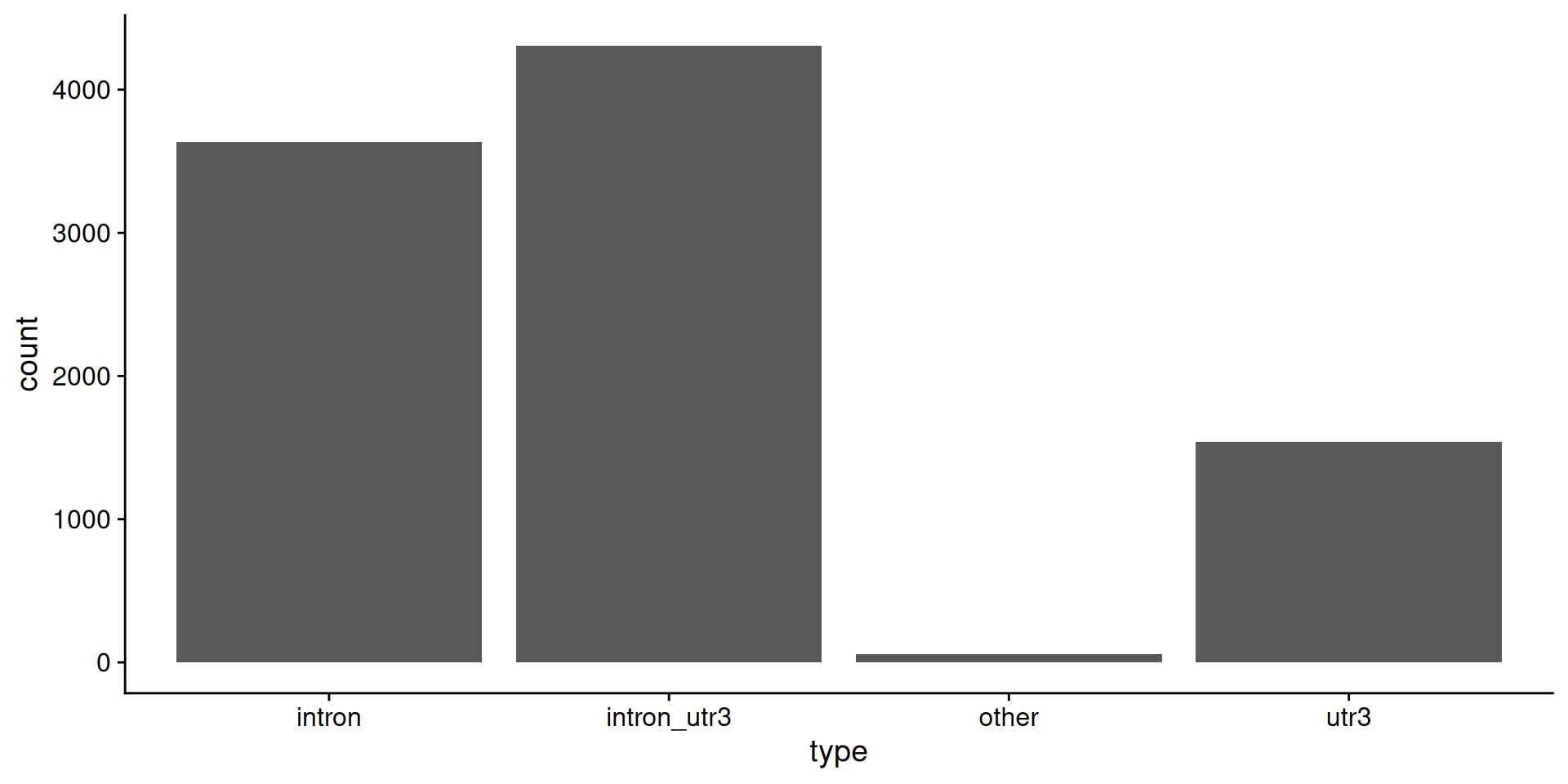
# let's keep 5' utr, cds, intron, 3' utr and intergenic
my_hg19_annots <- hg19_annots[c(3, 4, 7, 10, 11)]
# build the annotation database
annotations <- build_annotations(genome = "hg19", annotations = my_hg19_annots)'select()' returned 1:1 mapping between keys and
columnsBuilding promoters...Building 1to5kb upstream of TSS...Building intergenic...Building cds...Building 5UTRs...Building 3UTRs...Building exons...Building introns...GRanges object with 1446460 ranges and 5 metadata columns:
seqnames ranges strand |
<Rle> <IRanges> <Rle> |
[1] chr1 12190-12227 + |
[2] chr1 12595-12721 + |
[3] chr1 13403-13639 + |
[4] chr1 69091-70008 + |
[5] chr1 324343-324345 + |
... ... ... ... .
[1446456] chrUn_gl000246 1-38154 * |
[1446457] chrUn_gl000247 1-5786 * |
[1446458] chrUn_gl000247 10817-36422 * |
[1446459] chrUn_gl000248 1-39786 * |
[1446460] chrUn_gl000249 1-38502 * |
id tx_id gene_id
<character> <character> <character>
[1] CDS:1 uc010nxq.1 100287102
[2] CDS:2 uc010nxq.1 100287102
[3] CDS:3 uc010nxq.1 100287102
[4] CDS:4 uc001aal.1 79501
[5] CDS:5 uc009vjk.2 100133331
... ... ... ...
[1446456] intergenic:17023 <NA> <NA>
[1446457] intergenic:17024 <NA> <NA>
[1446458] intergenic:17025 <NA> <NA>
[1446459] intergenic:17026 <NA> <NA>
[1446460] intergenic:17027 <NA> <NA>
symbol type
<character> <character>
[1] DDX11L1 hg19_genes_cds
[2] DDX11L1 hg19_genes_cds
[3] DDX11L1 hg19_genes_cds
[4] OR4F5 hg19_genes_cds
[5] <NA> hg19_genes_cds
... ... ...
[1446456] <NA> hg19_genes_intergenic
[1446457] <NA> hg19_genes_intergenic
[1446458] <NA> hg19_genes_intergenic
[1446459] <NA> hg19_genes_intergenic
[1446460] <NA> hg19_genes_intergenic
-------
seqinfo: 93 sequences (1 circular) from hg19 genome
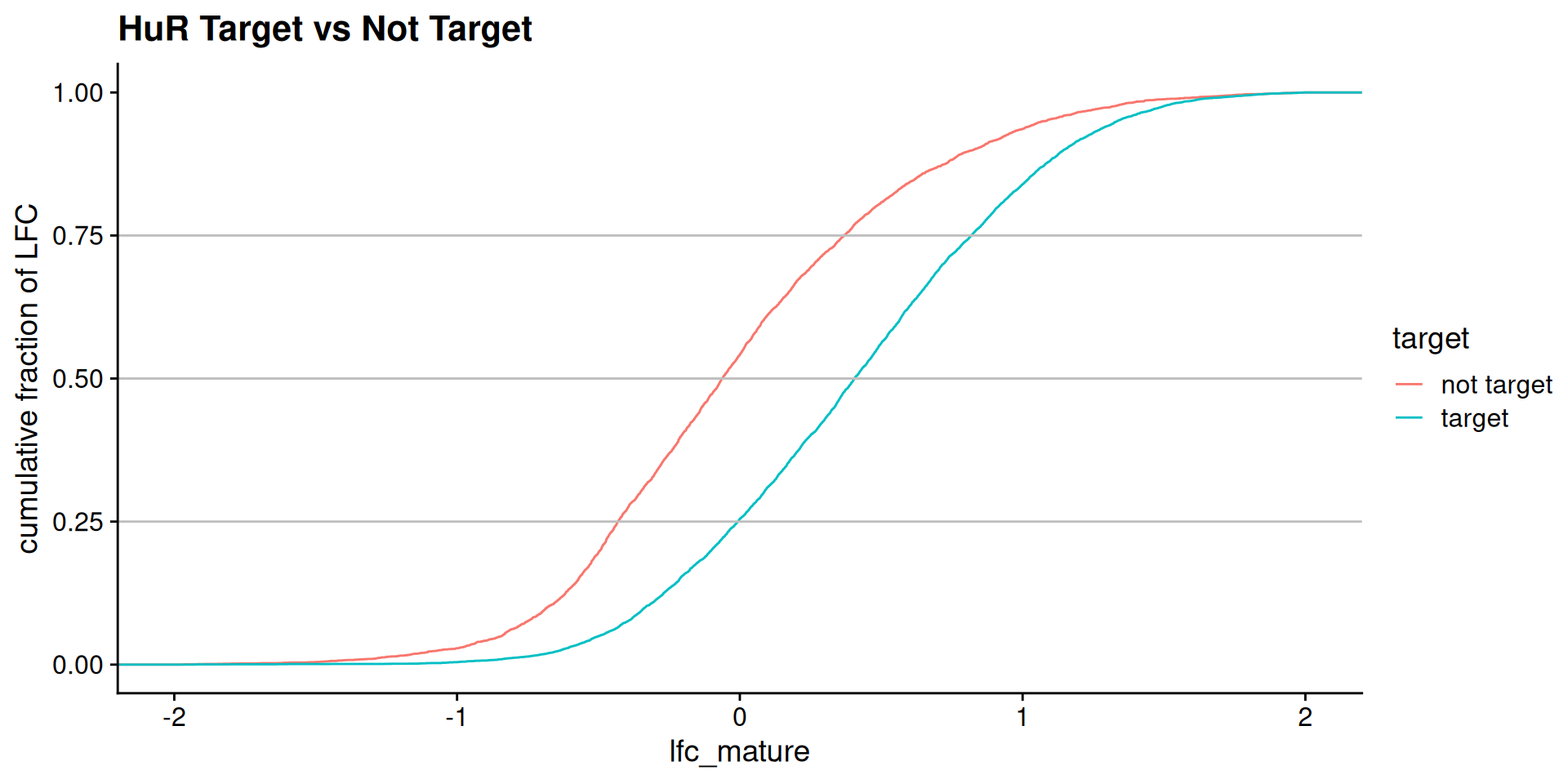
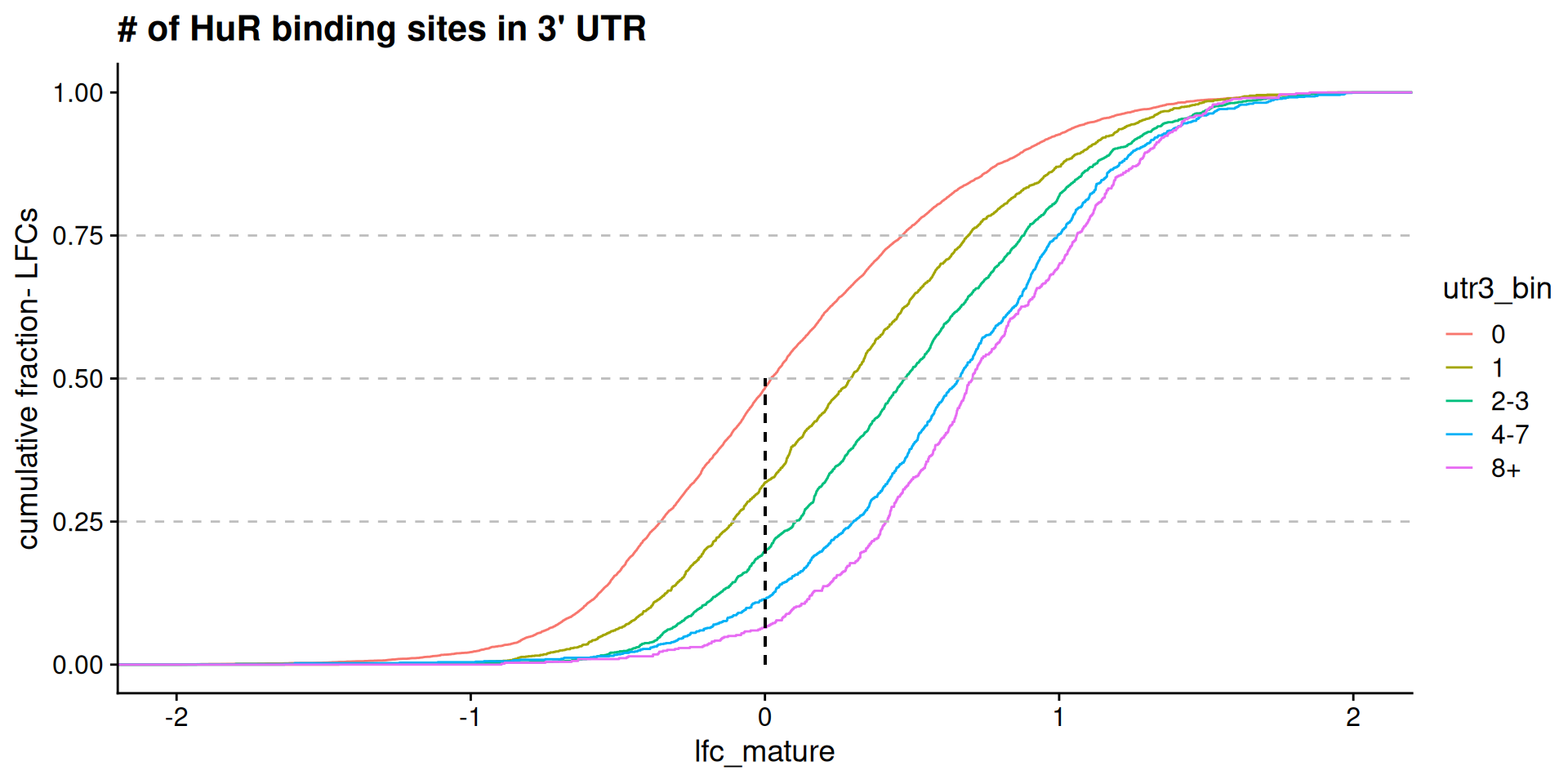
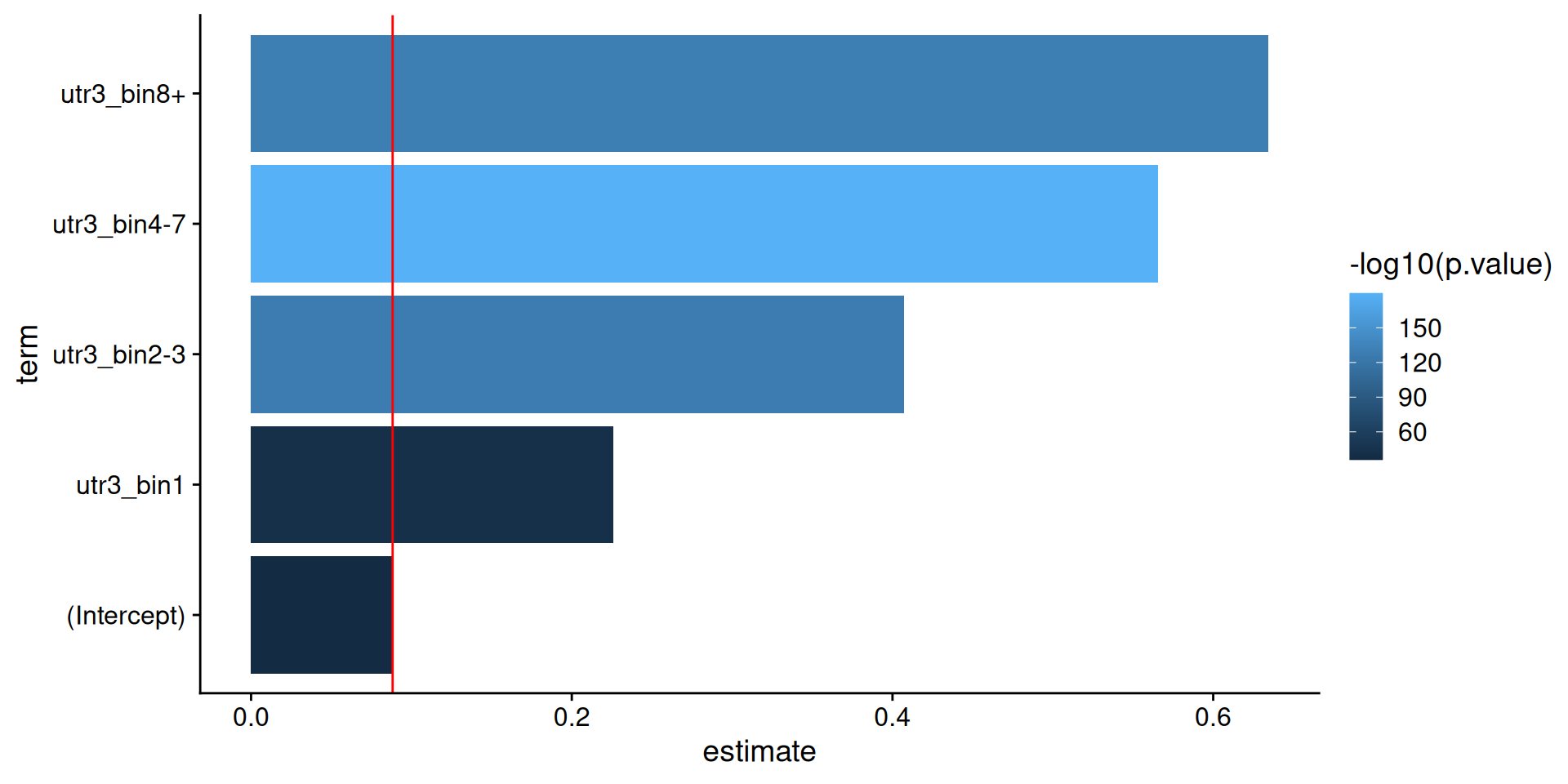
 Model predictions:
Model predictions: