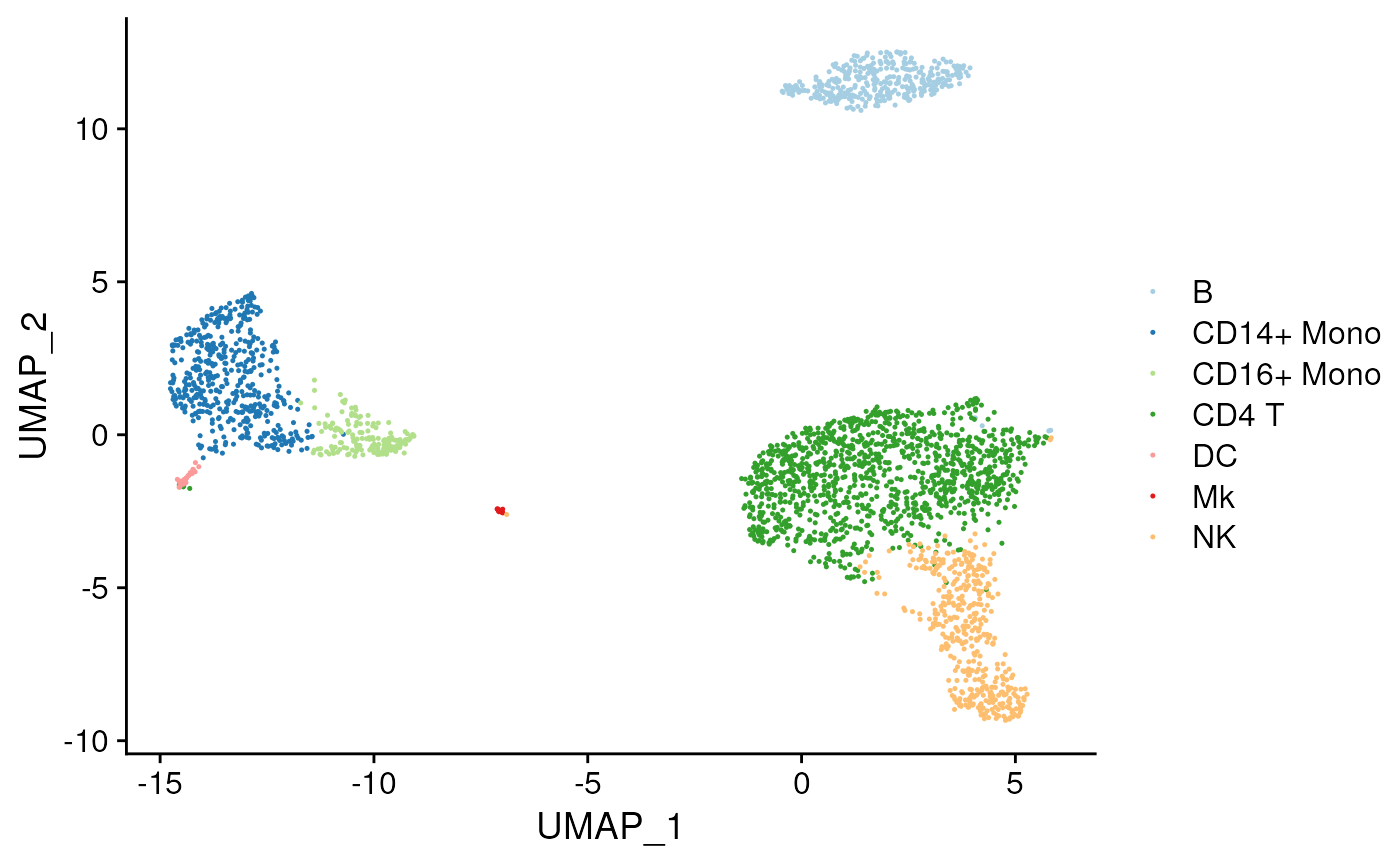
Plot best calls for each cluster on a tSNE or umap
Usage
plot_best_call(
cor_mat,
metadata,
cluster_col = "cluster",
collapse_to_cluster = FALSE,
threshold = 0,
x = "UMAP_1",
y = "UMAP_2",
plot_r = FALSE,
per_cell = FALSE,
...
)Arguments
- cor_mat
input similarity matrix
- metadata
input metadata with tsne or umap coordinates and cluster ids
- cluster_col
metadata column, can be cluster or cellid
- collapse_to_cluster
if a column name is provided, takes the most frequent call of entire cluster to color in plot
- threshold
minimum correlation coefficent cutoff for calling clusters
- x
x variable
- y
y variable
- plot_r
whether to include second plot of cor eff for best call
- per_cell
whether the cor_mat was generate per cell or per cluster
- ...
passed to plot_dims
Examples
res <- clustify(
input = pbmc_matrix_small,
metadata = pbmc_meta,
ref_mat = cbmc_ref,
query_genes = pbmc_vargenes,
cluster_col = "classified"
)
#> using # of genes: 599
#> similarity computation completed, matrix of 9 x 13, preparing output
plot_best_call(
cor_mat = res,
metadata = pbmc_meta,
cluster_col = "classified"
)